Instantly share code, notes, and snippets.

shantanuatgit / Assignment3.py
- Download ZIP
- Star 0 You must be signed in to star a gist
- Fork 0 You must be signed in to fork a gist
- Embed Embed this gist in your website.
- Share Copy sharable link for this gist.
- Clone via HTTPS Clone using the web URL.
- Learn more about clone URLs
- Save shantanuatgit/2054ad91d1b502bae4a8965d6fb297e1 to your computer and use it in GitHub Desktop.
Browse Course Material
Course info.
- Dr. Casey Rodriguez
Departments
- Mathematics
As Taught In
- Mathematical Analysis
Learning Resource Types
Real analysis, assignment 3 (pdf).

You are leaving MIT OpenCourseWare

Assignment 3: Hello Vectors
Assignment 3: hello vectors #.
Welcome to this week’s programming assignment of the specialization. In this assignment we will explore word vectors. In natural language processing, we represent each word as a vector consisting of numbers. The vector encodes the meaning of the word. These numbers (or weights) for each word are learned using various machine learning models, which we will explore in more detail later in this specialization. Rather than make you code the machine learning models from scratch, we will show you how to use them. In the real world, you can always load the trained word vectors, and you will almost never have to train them from scratch. In this assignment you will
Predict analogies between words.
Use PCA to reduce the dimensionality of the word embeddings and plot them in two dimensions.
Compare word embeddings by using a similarity measure (the cosine similarity).
Understand how these vector space models work.
1.0 Predict the Countries from Capitals #
During the presentation of the module, we have illustrated the word analogies by finding the capital of a country from the country. In this part of the assignment we have changed the problem a bit. You are asked to predict the countries that corresponde to some capitals . You are playing trivia against some second grader who just took their geography test and knows all the capitals by heart. Thanks to NLP, you will be able to answer the questions properly. In other words, you will write a program that can give you the country by its capital. That way you are pretty sure you will win the trivia game. We will start by exploring the data set.
Important Note on Submission to the AutoGrader #
Before submitting your assignment to the AutoGrader, please make sure you are not doing the following:
You have not added any extra print statement(s) in the assignment.
You have not added any extra code cell(s) in the assignment.
You have not changed any of the function parameters.
You are not using any global variables inside your graded exercises. Unless specifically instructed to do so, please refrain from it and use the local variables instead.
You are not changing the assignment code where it is not required, like creating extra variables.
If you do any of the following, you will get something like, Grader not found (or similarly unexpected) error upon submitting your assignment. Before asking for help/debugging the errors in your assignment, check for these first. If this is the case, and you don’t remember the changes you have made, you can get a fresh copy of the assignment by following these instructions .
1.1 Importing the data #
As usual, you start by importing some essential Python libraries and the load dataset. The dataset will be loaded as a Pandas DataFrame , which is very a common method in data science. Because of the large size of the data, this may take a few minutes.
To Run This Code On Your Own Machine: #
Note that because the original google news word embedding dataset is about 3.64 gigabytes, the workspace is not able to handle the full file set. So we’ve downloaded the full dataset, extracted a sample of the words that we’re going to analyze in this assignment, and saved it in a pickle file called word_embeddings_capitals.p
If you want to download the full dataset on your own and choose your own set of word embeddings, please see the instructions and some helper code.
Download the dataset from this page .
Search in the page for ‘GoogleNews-vectors-negative300.bin.gz’ and click the link to download.
You’ll need to unzip the file.
Copy-paste the code below and run it on your local machine after downloading the dataset to the same directory as the notebook.
Now we will load the word embeddings as a Python dictionary . As stated, these have already been obtained through a machine learning algorithm.
Each of the word embedding is a 300-dimensional vector.
Predict relationships among words #
Now you will write a function that will use the word embeddings to predict relationships among words.
The function will take as input three words.
The first two are related to each other.
It will predict a 4th word which is related to the third word in a similar manner as the two first words are related to each other.
As an example, “Athens is to Greece as Bangkok is to ______”?
You will write a program that is capable of finding the fourth word.
We will give you a hint to show you how to compute this.
A similar analogy would be the following:
You will implement a function that can tell you the capital of a country. You should use the same methodology shown in the figure above. To do this, you’ll first compute the cosine similarity metric or the Euclidean distance.
1.2 Cosine Similarity #
The cosine similarity function is:
\(A\) and \(B\) represent the word vectors and \(A_i\) or \(B_i\) represent index i of that vector. Note that if A and B are identical, you will get \(cos(\theta) = 1\) .
Otherwise, if they are the total opposite, meaning, \(A= -B\) , then you would get \(cos(\theta) = -1\) .
If you get \(cos(\theta) =0\) , that means that they are orthogonal (or perpendicular).
Numbers between 0 and 1 indicate a similarity score.
Numbers between -1 and 0 indicate a dissimilarity score.
Instructions : Implement a function that takes in two word vectors and computes the cosine distance.
- Python's NumPy library adds support for linear algebra operations (e.g., dot product, vector norm ...).
- Use numpy.dot .
- Use numpy.linalg.norm .
Expected Output :
\(\approx\) 0.651095
1.3 Euclidean distance #
You will now implement a function that computes the similarity between two vectors using the Euclidean distance. Euclidean distance is defined as:
\(n\) is the number of elements in the vector
\(A\) and \(B\) are the corresponding word vectors.
The more similar the words, the more likely the Euclidean distance will be close to 0.
Instructions : Write a function that computes the Euclidean distance between two vectors.
Expected Output:
1.4 Finding the country of each capital #
Now, you will use the previous functions to compute similarities between vectors, and use these to find the capital cities of countries. You will write a function that takes in three words, and the embeddings dictionary. Your task is to find the capital cities. For example, given the following words:
1: Athens 2: Greece 3: Baghdad,
your task is to predict the country 4: Iraq.
Instructions :
To predict the capital you might want to look at the King - Man + Woman = Queen example above, and implement that scheme into a mathematical function, using the word embeddings and a similarity function.
Iterate over the embeddings dictionary and compute the cosine similarity score between your vector and the current word embedding.
You should add a check to make sure that the word you return is not any of the words that you fed into your function. Return the one with the highest score.
Expected Output: (Approximately)
(‘Egypt’, 0.7626821)
1.5 Model Accuracy #
Now you will test your new function on the dataset and check the accuracy of the model:
Instructions : Write a program that can compute the accuracy on the dataset provided for you. You have to iterate over every row to get the corresponding words and feed them into you get_country function above.
- Use pandas.DataFrame.iterrows .
NOTE: The cell below takes about 30 SECONDS to run.
\(\approx\) 0.92
3.0 Plotting the vectors using PCA #
Now you will explore the distance between word vectors after reducing their dimension. The technique we will employ is known as principal component analysis (PCA) . As we saw, we are working in a 300-dimensional space in this case. Although from a computational perspective we were able to perform a good job, it is impossible to visualize results in such high dimensional spaces.
You can think of PCA as a method that projects our vectors in a space of reduced dimension, while keeping the maximum information about the original vectors in their reduced counterparts. In this case, by maximum infomation we mean that the Euclidean distance between the original vectors and their projected siblings is minimal. Hence vectors that were originally close in the embeddings dictionary, will produce lower dimensional vectors that are still close to each other.
You will see that when you map out the words, similar words will be clustered next to each other. For example, the words ‘sad’, ‘happy’, ‘joyful’ all describe emotion and are supposed to be near each other when plotted. The words: ‘oil’, ‘gas’, and ‘petroleum’ all describe natural resources. Words like ‘city’, ‘village’, ‘town’ could be seen as synonyms and describe a similar thing.
Before plotting the words, you need to first be able to reduce each word vector with PCA into 2 dimensions and then plot it. The steps to compute PCA are as follows:
Mean normalize the data
Compute the covariance matrix of your data ( \(\Sigma\) ).
Compute the eigenvectors and the eigenvalues of your covariance matrix
Multiply the first K eigenvectors by your normalized data. The transformation should look something as follows:
You will write a program that takes in a data set where each row corresponds to a word vector.
The word vectors are of dimension 300.
Use PCA to change the 300 dimensions to n_components dimensions.
The new matrix should be of dimension m, n_componentns .
First de-mean the data
Get the eigenvalues using linalg.eigh . Use ‘eigh’ rather than ‘eig’ since R is symmetric. The performance gain when using eigh instead of eig is substantial.
Sort the eigenvectors and eigenvalues by decreasing order of the eigenvalues.
Get a subset of the eigenvectors (choose how many principle components you want to use using n_components).
Return the new transformation of the data by multiplying the eigenvectors with the original data.
- Use numpy.mean(a,axis=None) : If you set axis = 0 , you take the mean for each column. If you set axis = 1 , you take the mean for each row. Remember that each row is a word vector, and the number of columns are the number of dimensions in a word vector.
- Use numpy.cov(m, rowvar=True) . This calculates the covariance matrix. By default rowvar is True . From the documentation: "If rowvar is True (default), then each row represents a variable, with observations in the columns." In our case, each row is a word vector observation, and each column is a feature (variable).
- Use numpy.linalg.eigh(a, UPLO='L')
- Use numpy.argsort sorts the values in an array from smallest to largest, then returns the indices from this sort.
- In order to reverse the order of a list, you can use: x[::-1] .
- To apply the sorted indices to eigenvalues, you can use this format x[indices_sorted] .
- When applying the sorted indices to eigen vectors, note that each column represents an eigenvector. In order to preserve the rows but sort on the columns, you can use this format x[:,indices_sorted]
- To transform the data using a subset of the most relevant principle components, take the matrix multiplication of the eigenvectors with the original data.
- The data is of shape (n_observations, n_features) .
- The subset of eigenvectors are in a matrix of shape (n_features, n_components) .
- To multiply these together, take the transposes of both the eigenvectors (n_components, n_features) and the data (n_features, n_observations).
- The product of these two has dimensions (n_components,n_observations) . Take its transpose to get the shape (n_observations, n_components) .
Your original matrix was: (3,10) and it became:
Now you will use your pca function to plot a few words we have chosen for you. You will see that similar words tend to be clustered near each other. Sometimes, even antonyms tend to be clustered near each other. Antonyms describe the same thing but just tend to be on the other end of the scale They are usually found in the same location of a sentence, have the same parts of speech, and thus when learning the word vectors, you end up getting similar weights. In the next week we will go over how you learn them, but for now let’s just enjoy using them.
Instructions: Run the cell below.
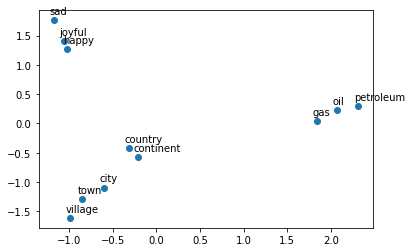
What do you notice?
The word vectors for gas, oil and petroleum appear related to each other, because their vectors are close to each other. Similarly, sad, joyful and happy all express emotions, and are also near each other.
Assignment 3: SAT Problems Solving
In logic or computer science, the Boolean Satisfiability Problem (abbreviated as SAT in this assignment) is to determine whether or not a given propositional logic formulae is true, and to further determine the model which makes the formulae true. The program or tool to answer the SAT problem is called an SAT solver. In this assignment, we'll learn how a SAT solver works and how to encode propositions and check its satisfiability by such SAT solvers. And as applications, we will also learn how to model practical problems by satisfiability, and thus solve them with the aid of SAT solvers.
There are many SAT/SMT solvers available, each with its pros and cons. The solver we'll be using in this assignment is the Z3 theorem solver/prover , developed by Microsoft Research. There is no special reason for us to choose Z3, any other SAT solvers will also be OK, but Z3's Python APIs will be very convenient.
This assignment is divided into four parts, each of which contains both some tutorials and problems. The first part is the SAT encoding of the basic propositions; the second part covers validity checking; part three covers the DPLL algorithm implementation; and the fourth part covers some SAT applications. Some problems are tagged with Exercise , which you should solve. And several problems are tagged with Challenge , which are optional.
Before starting with this assignment, make sure you've finished Software Setup in the assignment 1, and have Z3 and Python properly installed on your computer. For any installation problems, please feel free to contact us for help. As we'll be using Z3's Python-based API; you may find it useful to refer to the z3py tutorial and the documentation for Z3's Python API .
Hand-in Procedure
When you finished the assignment, zip you code files with file name studentid-assignment3.zip (e.g SA19225111-assignment3.zip ), and submit it to Postgraduate Information Platform . The deadline TBA (Beijing time). Any late submission will NOT be accepted.
Part A: Basic Propositional Logic
In this section, let's start by learning how to solve basic propositional logic problems with Z3.
Declare propositions:
In Z3, we can declare two propositions to be just booleans, this is rather natural, for propositions can have values true or false. To declare two propositions P and Q :
Or, we can use a more compact shorthand:
Build propositions with connectives:
Z3 supports connectives: /\(And) , \/(Or) , ~(Not) , ->(Implies) , along with several others. We can build propositions by writing Lisp-style abstract syntax trees directly, for example, the disjunction: P \/ Q (Note that the connective \/ is called Or in Z3) can be encoded as the following AST:
Usage of Solve:
Example A-1: The simplest usage for Z3 is to feed the proposition to Z3 directly, to check the satisfiability, this can be done by calling the solve() function, the solve() function will create an instance of solver, check the satisfiability of the proposition, and output a model if that proposition is satisfiable, the code looks like:
For the above call, Z3 will output something like this:
which is a model with assignments to proposition P and Q that makes the proposition F satisfiable. Obviously, this is just one of several possible models.
Example A-2: Not all propositions are satisfiable, consider this proposition:
Z3 will output:
Obtain More Possible Solutions:
Consider again example A-1 in the previous:
After all, we're asking Z3 about the satisfiability of the proposition, so one row of evidence is enough. What if we want Z3 to output all the assignment of propositions that make the proposition satisfiable, not just the first one? For the above example, we want the all first 3 rows. Here is the trick: when we get an answer, we negate the answer, make a conjunction with the original proposition, then check the satisfiability again. For the above example:
The output will obtained all the 3 possible solutions:
Part B: SAT And Validity
In Exercise 1, we've learned how to represent propositions in Z3 and how to use Z3 to obtain the solutions that make a given proposition satisfiable. In this part, we continue to discuss how to use Z3 to check the validity of propositions. Recall in exercise 2, we once used theorem prover to prove the validity of propositions, so this is another strategy.
Example B-1: As we've discussed in previous lecture, the relationship between the validity and satisfiability of a proposition P is established by: valid(P) unsat(~P) Let consider our previous example:
Z3 will output the following solution: [P = False, Q = False] the fact that ~F is satisfiable means that the proposition F is not valid. By this, it should be very clear how to use solvers like Z3 to prove the validity of a proposition.
Example B-2: Now we try to prove the double negation law( ~~P -> P ) is valid:
Part C: The DPLL algorithm
- nnf(P) : to convert the proposition P into negation normal form;
- cnf(P) : to convert the proposition P into conjunction normal form;
- dpll(P) : to calculate the satisfiability P of the proposition P ;
After finishing this algorithm to experiment how large propositions your algorithm can solve. For instance, you can generate some very large propositions using this generator and feed the generated propositions to your solver.
How to make you DPLL more efficient, one idea to make your solver concurrent. To be specific, for the splitting step, to the two cases, instead of using two sequential calls, we can create two threads/processes to do concurrent calls.
Part D: Applications
In the previous part we've discussed how to obtain solutions and prove the validity for propositions, and implemented the DPLL algorithm. In this part, we will try to use Z3 to solve some practical problems.
Usually when engineers design circuit layouts, they need do some verifications to make sure those layouts will not only output a permanent electrical signal since it's useless. We want to guarantee that the layouts can output different signals based on the inputs.

As the graph above shows, there are three kinds of logic gates used in design circuit layout, And, Or, Not. And there four inputs in the graph, A, B, C and D.
- Alice can not sit near to Carol;
- Bob can not sit right to Alice.
- Is there any possible arrangement?
- How many possible different arrangement in total?

- one person can only takes just one seat;
- one seat can only be taken by one person;

- each row has just 1 queen;
- each column has just 1 queen;
- each diagonal has at most 1 queen;
- each anti-diagonal has at most 1 queen;
- How long does your program take to solve 8-queen?
- How long does your program take to solve 100-queens?
- What's the maximal of N your program can solve?
Happy hacking!
API for Assignments 3.1 and 3.2
This page describes the endpoints for the API you will use as a client in assignment 3.1 and implement in assignment 3.2.
GET /users/:id
Post /users, patch /users/:id, get /users/:id/feed, post /users/:id/posts, post /users/:id/follow, delete /users/:id/follow, general notes.
- All endpoints return JSON. The response will always be an object (so e.g. if an endpoint returns an array, it will be wrapped in an object with a single key/value pair).
- When the endpoint returns an error status, the response body will be an object with an error key, whose value is a human-readable error message.
- In the examples, lines that begin with > are the request, and lines with no prefix are the response.
- Unless stated explicitly, elements in an array or key/value pairs in an object are in no particular order.
An endpoint to confirm the API is running. You won't have to use it, but it may be useful for testing. For assignment 3.2, it is already implemented.
Returns: An object with basic info about the database.
Get a list of existing users.
Returns: An object with a single key, users , mapping to an array of user IDs.
Get the profile for the user id .
Returns: An object with the keys id , name , avatarURL (which are all strings), and following (which is an array of user IDs the given user is following).
- 404 if the user does not exist
Create a new user. The new user's name is the same as their ID, their avatar URL is the default avatar ( images/default.png ), and they are not following anyone.
Request body: An object with a single key id , whose value is the new user's ID.
Returns: Same as GET /users/:id after user is created.
- 400 if the request body is missing an id property, or the id is empty
- 400 if the user already exists
Update a user's profile (name or avatar URL). User IDs cannot be changed, but including the id in the request body is allowed.
If the specified display name is the empty string, the user's display name is reset to their user ID. If the specified avatar URL is the empty string, the user's avatar is reset to the default avatar ( images/default.png ).
Request body: An object with any number of the keys id (ignored), name , and avatarURL .
Note: Explicitly sending an empty string for name / avatarURL will set them to their defaults; not including them at all will leave them unchanged.
Returns: Same as GET /users/:id after user is updated.
Note: For assignment 3.1, our API checks if extra keys are included in the request body and returns an error if so. For assignment 3.2, you do not have to check for this case.
Returns: An object with a single key posts , mapping to an array. Each element has the following structure:
Create a new post by the given user. The post time is set to the current date/time.
Request body: An object with a single key text , containing the post text.
Returns: The exact object { "success": true } .
- 400 if the request body is missing a text property, or the text is empty
Have the user id follow the target user.
Query string: target : the ID of the user to follow
- 404 if either user id or target does not exist
- 400 if the query string is missing a target property, or the target is empty
- 400 if the user is already following the target
- 400 if the requesting user is the same as the target
Have the user id stop following the target user.
Query string: target : the ID of the user to stop following
- 404 if the user id does not exist
- 400 if the target user isn't being followed by the requesting user
Group Admins
- Show: — Everything — Updates Group Memberships Group Updates Topics Replies
- WordPress.org
- Documentation
- Learn WordPress
- Open access
- Published: 15 April 2024
Demuxafy : improvement in droplet assignment by integrating multiple single-cell demultiplexing and doublet detection methods
- Drew Neavin 1 ,
- Anne Senabouth 1 ,
- Himanshi Arora 1 , 2 ,
- Jimmy Tsz Hang Lee 3 ,
- Aida Ripoll-Cladellas 4 ,
- sc-eQTLGen Consortium ,
- Lude Franke 5 ,
- Shyam Prabhakar 6 , 7 , 8 ,
- Chun Jimmie Ye 9 , 10 , 11 , 12 ,
- Davis J. McCarthy 13 , 14 ,
- Marta Melé 4 ,
- Martin Hemberg 15 &
- Joseph E. Powell ORCID: orcid.org/0000-0002-5070-4124 1 , 16
Genome Biology volume 25 , Article number: 94 ( 2024 ) Cite this article
Metrics details
Recent innovations in single-cell RNA-sequencing (scRNA-seq) provide the technology to investigate biological questions at cellular resolution. Pooling cells from multiple individuals has become a common strategy, and droplets can subsequently be assigned to a specific individual by leveraging their inherent genetic differences. An implicit challenge with scRNA-seq is the occurrence of doublets—droplets containing two or more cells. We develop Demuxafy, a framework to enhance donor assignment and doublet removal through the consensus intersection of multiple demultiplexing and doublet detecting methods. Demuxafy significantly improves droplet assignment by separating singlets from doublets and classifying the correct individual.
Droplet-based single-cell RNA sequencing (scRNA-seq) technologies have provided the tools to profile tens of thousands of single-cell transcriptomes simultaneously [ 1 ]. With these technological advances, combining cells from multiple samples in a single capture is common, increasing the sample size while simultaneously reducing batch effects, cost, and time. In addition, following cell capture and sequencing, the droplets can be demultiplexed—each droplet accurately assigned to each individual in the pool [ 2 , 3 , 4 , 5 , 6 , 7 ].
Many scRNA-seq experiments now capture upwards of 20,000 droplets, resulting in ~16% (3,200) doublets [ 8 ]. Current demultiplexing methods can also identify doublets—droplets containing two or more cells—from different individuals (heterogenic doublets). These doublets can significantly alter scientific conclusions if they are not effectively removed. Therefore, it is essential to remove doublets from droplet-based single-cell captures.
However, demultiplexing methods cannot identify droplets containing multiple cells from the same individual (homogenic doublets) and, therefore, cannot identify all doublets in a single capture. If left in the dataset, those doublets could appear as transitional cells between two distinct cell types or a completely new cell type. Accordingly, additional methods have been developed to identify heterotypic doublets (droplets that contain two cells from different cell types) by comparing the transcriptional profile of each droplet to doublets simulated from the dataset [ 9 , 10 , 11 , 12 , 13 , 14 , 15 ]. It is important to recognise that demultiplexing methods achieve two functions—segregation of cells from different donors and separation of singlets from doublets—while doublet detecting methods solely classify singlets versus doublets.
Therefore, demultiplexing and transcription-based doublet detecting methods provide complementary information to improve doublet detection, providing a cleaner dataset and more robust scientific results. There are currently five genetic-based demultiplexing [ 2 , 3 , 4 , 5 , 6 , 7 , 16 ] and seven transcription-based doublet-detecting methods implemented in various languages [ 9 , 10 , 11 , 12 , 13 , 14 , 15 ]. Under different scenarios, each method is subject to varying performance and, in some instances, biases in their ability to accurately assign cells or detect doublets from certain conditions. The best combination of methods is currently unclear but will undoubtedly depend on the dataset and research question.
Therefore, we set out to identify the best combination of genetic-based demultiplexing and transcription-based doublet-detecting methods to remove doublets and partition singlets from different donors correctly. In addition, we have developed a software platform ( Demuxafy ) that performs these intersectional methods and provides additional commands to simplify the execution and interpretation of results for each method (Fig. 1 a).
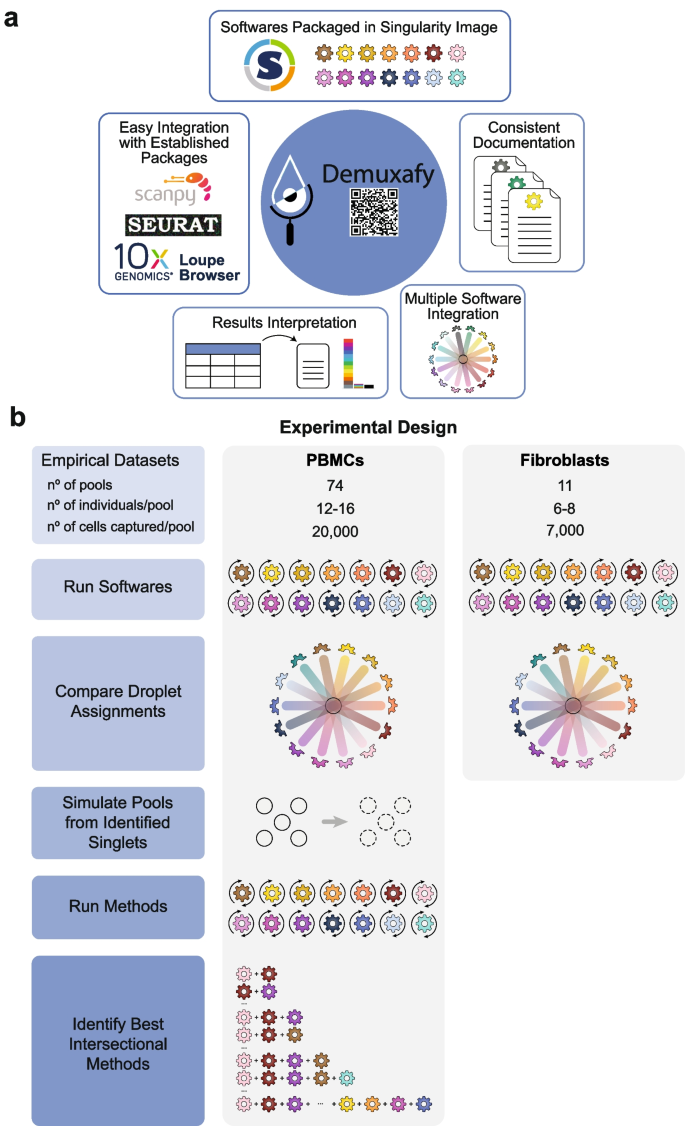
Study design and qualitative method classifications. a Demuxafy is a platform to perform demultiplexing and doublet detecting with consistent documentation. Demuxafy also provides wrapper scripts to quickly summarize the results from each method and assign clusters to each individual with reference genotypes when a reference-free demultiplexing method is used. Finally, Demuxafy provides a script to easily combine the results from multiple different methods into a single data frame and it provides a final assignment for each droplet based on the combination of multiple methods. In addition, Demuxafy provides summaries of the number of droplets classified as singlets or doublets by each method and a summary of the number of droplets assigned to each individual by each of the demultiplexing methods. b Two datasets are included in this analysis - a PBMC dataset and a fibroblast dataset. The PBMC dataset contains 74 pools that captured approximately 20,000 droplets each with 12-16 donor cells multiplexed per pool. The fibroblast dataset contains 11 pools of roughly 7,000 droplets per pool with sizes ranging from six to eight donors per pool. All pools were processed by all demultiplexing and doublet detecting methods and the droplet and donor classifications were compared between the methods and between the PBMCs and fibroblasts. Then the PBMC droplets that were classified as singlets by all methods were taken as ‘true singlets’ and used to generate new pools in silico. Those pools were then processed by each of the demultiplexing and doublet detecting methods and intersectional combinations of demultiplexing and doublet detecting methods were tested for different experimental designs
To compare the demultiplexing and doublet detecting methods, we utilised two large, multiplexed datasets—one that contained ~1.4 million peripheral blood mononuclear cells (PBMCs) from 1,034 donors [ 17 ] and one with ~94,000 fibroblasts from 81 donors [ 18 ]. We used the true singlets from the PBMC dataset to generate new in silico pools to assess the performance of each method and the multi-method intersectional combinations (Fig. 1 b).
Here, we compare 14 demultiplexing and doublet detecting methods with different methodological approaches, capabilities, and intersectional combinations. Seven of those are demultiplexing methods ( Demuxalot [ 6 ], Demuxlet [ 3 ], Dropulation [ 5 ], Freemuxlet [ 16 ], ScSplit [ 7 ], Souporcell [ 4 ], and Vireo [ 2 ]) which leverage the common genetic variation between individuals to identify cells that came from each individual and to identify heterogenic doublets. The seven remaining methods ( DoubletDecon [ 9 ], DoubletDetection [ 14 ], DoubletFinder [ 10 ], ScDblFinder [ 11 ], Scds [ 12 ], Scrublet [ 13 ], and Solo [ 15 ]) identify doublets based on their similarity to simulated doublets generated by adding the transcriptional profiles of two randomly selected droplets in the dataset. These methods assume that the proportion of real doublets in the dataset is low, so combining any two droplets will likely represent the combination of two singlets.
We identify critical differences in the performance of demultiplexing and doublet detecting methods to classify droplets correctly. In the case of the demultiplexing techniques, their performance depends on their ability to identify singlets from doublets and assign a singlet to the correct individual. For doublet detecting methods, the performance is based solely on their ability to differentiate a singlet from a doublet. We identify limitations in identifying specific doublet types and cell types by some methods. In addition, we compare the intersectional combinations of these methods for multiple experimental designs and demonstrate that intersectional approaches significantly outperform all individual techniques. Thus, the intersectional methods provide enhanced singlet classification and doublet removal—a critical but often under-valued step of droplet-based scRNA-seq processing. Our results demonstrate that intersectional combinations of demultiplexing and doublet detecting software provide significant advantages in droplet-based scRNA-seq preprocessing that can alter results and conclusions drawn from the data. Finally, to provide easy implementation of our intersectional approach, we provide Demuxafy ( https://demultiplexing-doublet-detecting-docs.readthedocs.io/en/latest/index.html ) a complete platform to perform demultiplexing and doublet detecting intersectional methods (Fig. 1 a).
Study design
To evaluate demultiplexing and doublet detecting methods, we developed an experimental design that applies the different techniques to empirical pools and pools generated in silico from the combination of true singlets—droplets identified as singlets by every method (Fig. 1 a). For the first phase of this study, we used two empirical multiplexed datasets—the peripheral blood mononuclear cell (PBMC) dataset containing ~1.4 million cells from 1034 donors and a fibroblast dataset of ~94,000 cells from 81 individuals (Additional file 1 : Table S1). We chose these two cell systems to assess the methods in heterogeneous (PBMC) and homogeneous (fibroblast) cell types.
Demultiplexing and doublet detecting methods perform similarly for heterogeneous and homogeneous cell types
We applied the demultiplexing methods ( Demuxalot , Demuxlet , Dropulation , Freemuxlet , ScSplit , Souporcell , and Vireo ) and doublet detecting methods ( DoubletDecon , DoubletDetection , DoubletFinder , ScDblFinder , Scds , Scrublet , and Solo ) to the two datasets and assessed the results from each method. We first compared the droplet assignments by identifying the number of singlets and doublets identified by a given method that were consistently annotated by all methods (Fig. 2 a–d). We also identified the percentage of droplets that were annotated consistently between pairs of methods (Additional file 2 : Fig S1). In the cases where two demultiplexing methods were compared to one another, both the droplet type (singlet or doublet) and the assignment of the droplet to an individual had to match to be considered in agreement. In all other comparisons (i.e. demultiplexing versus doublet detecting and doublet detecting versus doublet detecting), only the droplet type (singlet or doublet) was considered for agreement since doublet detecting methods cannot annotate donor assignment. We found that the two method types were more similar to other methods of the same type (i.e., demultiplexing versus demultiplexing and doublet detecting versus doublet detecting) than they were to methods from a different type (demultiplexing methods versus doublet detecting methods; Supplementary Fig 1). We found that the similarity of the demultiplexing and doublet detecting methods was consistent in the PBMC and fibroblast datasets (Pearson correlation R = 0.78, P -value < 2×10 −16 ; Fig S1a-c). In addition, demultiplexing methods were more similar than doublet detecting methods for both the PBMC and fibroblast datasets (Wilcoxon rank-sum test: P < 0.01; Fig. 2 a–b and Additional file 2 : Fig S1).
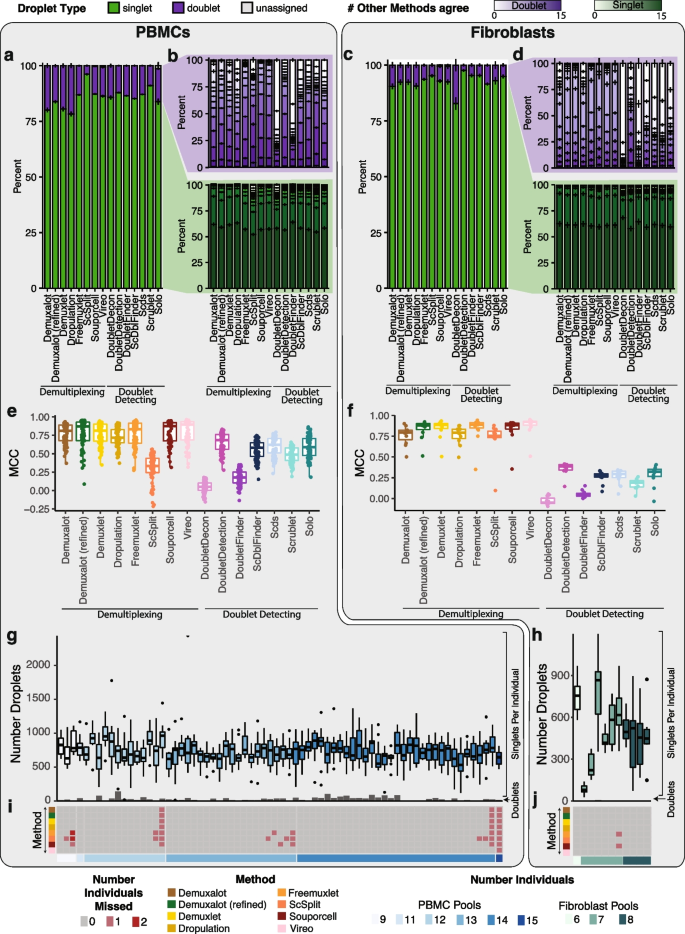
Demultiplexing and Doublet Detecting Method Performance Comparison. a The proportion of droplets classified as singlets and doublets by each method in the PBMCs. b The number of other methods that classified the singlets and doublets identified by each method in the PBMCs. c The proportion of droplets classified as singlets and doublets by each method in the fibroblasts. d The number of other methods that classified the singlets and doublets identified by each method in the fibroblasts. e - f The performance of each method when the majority classification of each droplet is considered the correct annotation in the PBMCs ( e ) and fibroblasts ( f ). g - h The number of droplets classified as singlets (box plots) and doublets (bar plots) by all methods in the PBMC ( g ) and fibroblast ( h ) pools. i - j The number of donors that were not identified by each method in each pool for PBMCs ( i ) and fibroblasts ( j ). PBMC: peripheral blood mononuclear cell. MCC: Matthew’s correlationcoefficient
The number of unique molecular identifiers (UMIs) and genes decreased in droplets that were classified as singlets by a larger number of methods while the mitochondrial percentage increased in both PBMCs and fibroblasts (Additional file 2 : Fig S2).
We next interrogated the performance of each method using the Matthew’s correlation coefficient (MCC) to calculate the consistency between Demuxify and true droplet classification. We identified consistent trends in the MCC scores for each method between the PBMCs (Fig. 2 e) and fibroblasts (Fig. 2 f). These data indicate that the methods behave similarly, relative to one another, for heterogeneous and homogeneous datasets.
Next, we sought to identify the droplets concordantly classified by all demultiplexing and doublet detecting methods in the PBMC and fibroblast datasets. On average, 732 singlets were identified for each individual by all the methods in the PBMC dataset. Likewise, 494 droplets were identified as singlets for each individual by all the methods in the fibroblast pools. However, the concordance of doublets identified by all methods was very low for both datasets (Fig. 2 e–f). Notably, the consistency of classifying a droplet as a doublet by all methods was relatively low (Fig. 2 b,d,g, and h). This suggests that doublet identification is not consistent between all the methods. Therefore, further investigation is required to identify the reasons for these inconsistencies between methods. It also suggests that combining multiple methods for doublet classification may be necessary for more complete doublet removal. Further, some methods could not identify all the individuals in each pool (Fig. 2 i–j). The non-concordance between different methods demonstrates the need to effectively test each method on a dataset where the droplet types are known.
Computational resources vary for demultiplexing and doublet detecting methods
We recorded each method’s computational resources for the PBMC pools, with ~20,000 cells captured per pool (Additional file 1 : Table S1). Of the demultiplexing methods, ScSplit took the most time (multiple days) and required the most steps, but Demuxalot , Demuxlet , and Freemuxlet used the most memory. Solo took the longest time (median 13 h) and most memory to run for the doublet detecting methods but is the only method built to be run directly from the command line, making it easy to implement (Additional file 2 : Fig S3).
Generate pools with known singlets and doublets
However, there is no gold standard to identify which droplets are singlets or doublets. Therefore, in the second phase of our experimental design (Fig. 1 b), we used the PBMC droplets classified as singlets by all methods to generate new pools in silico. We chose to use the PBMC dataset since our first analyses indicated that method performance is similar for homogeneous (fibroblast) and heterogeneous (PBMC) cell types (Fig. 2 and Additional file 2 : Fig S1) and because we had many more individuals available to generate in silico pools from the PBMC dataset (Additional file 1 : Table S1).
We generated 70 pools—10 each of pools that included 2, 4, 8, 16, 32, 64, or 128 individuals (Additional file 1 : Table S2). We assume a maximum 20% doublet rate as it is unlikely researchers would use a technology that has a higher doublet rate (Fig. 3 a).
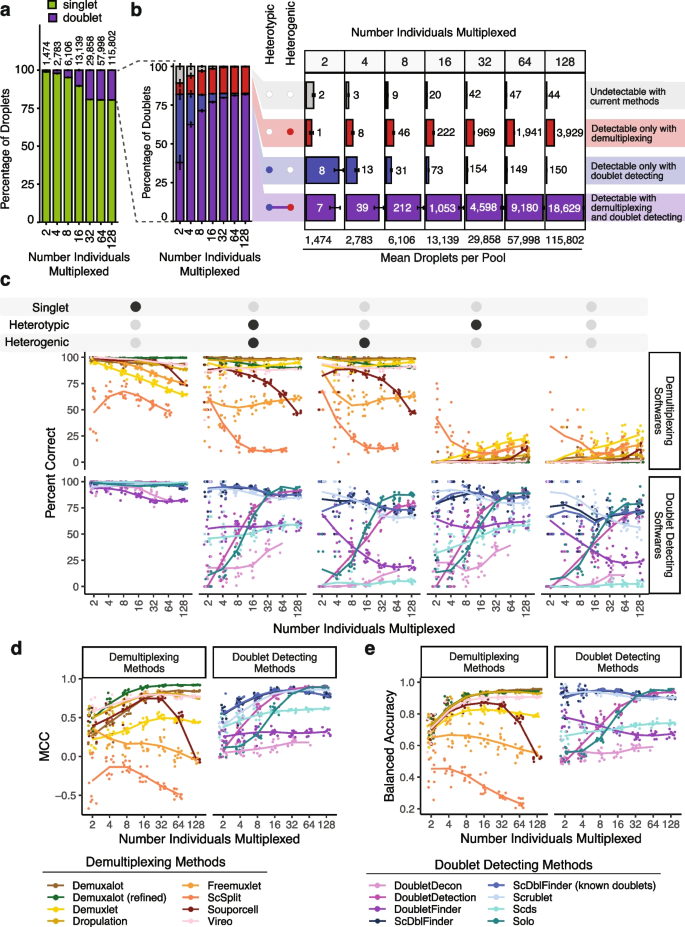
In silico Pool Doublet Annotation and Method Performance. a The percent of singlets and doublets in the in -silico pools - separated by the number of multiplexed individuals per pool. b The percentage and number of doublets that are heterogenic (detectable by demultiplexing methods), heterotypic (detectable by doublet detecting methods), both (detectable by either method category) and neither (not detectable with current methods) for each multiplexed pool size. c Percent of droplets that each of the demultiplexing and doublet detecting methods classified correctly for singlets and doublet subtypes for different multiplexed pool sizes. d Matthew’s Correlation Coefficient (MCC) for each of the methods for each of the multiplexed pool sizes. e Balanced accuracy for each of the methods for each of the multiplexed pool sizes
We used azimuth to classify the PBMC cell types for each droplet used to generate the in silico pools [ 19 ] (Additional file 2 : Fig S4). As these pools have been generated in silico using empirical singlets that have been well annotated, we next identified the proportion of doublets in each pool that were heterogenic, heterotypic, both, and neither. This approach demonstrates that a significant percentage of doublets are only detectable by doublet detecting methods (homogenic and heterotypic) for pools with 16 or fewer donors multiplexed (Fig. 3 b).
While the total number of doublets that would be missed if only using demultiplexing methods appears small for fewer multiplexed individuals (Fig. 3 b), it is important to recognise that this is partly a function of the ~732 singlet cells per individual used to generate these pools. Hence, the in silico pools with fewer individuals also have fewer cells. Therefore, to obtain numbers of doublets that are directly comparable to one another, we calculated the number of each doublet type that would be expected to be captured with 20,000 cells when 2, 4, 8, 16, or 32 individuals were multiplexed (Additional file 2 : Fig S5). These results demonstrate that many doublets would be falsely classified as singlets since they are homogenic when just using demultiplexing methods for a pool of 20,000 cells captured with a 16% doublet rate (Additional file 2 : Fig S5). However, as more individuals are multiplexed, the number of droplets that would not be detectable by demultiplexing methods (homogenic) decreases. This suggests that typical workflows that use only one demultiplexing method to remove doublets from pools that capture 20,000 droplets with 16 or fewer multiplexed individuals fail to adequately remove between 173 (16 multiplexed individuals) and 1,325 (2 multiplexed individuals) doublets that are homogenic and heterotypic which could be detected by doublet detecting methods (Additional file 2 : Fig S5). Therefore, a technique that uses both demultiplexing and doublet detecting methods in parallel will complement more complete doublet removal methods. Consequently, we next set out to identify the demultiplexing and doublet detecting methods that perform the best on their own and in concert with other methods.
Doublet and singlet droplet classification effectiveness varies for demultiplexing and doublet detecting methods
Demultiplexing methods fail to classify homogenic doublets.
We next investigated the percentage of the droplets that were correctly classified by each demultiplexing and doublet detecting method. In addition to the seven demultiplexing methods, we also included Demuxalot with the additional steps to refine the genotypes that can then be used for demultiplexing— Demuxalot (refined). Demultiplexing methods correctly classify a large portion of the singlets and heterogenic doublets (Fig. 3 c). This pattern is highly consistent across different cell types, with the notable exceptions being decreased correct classifications for erythrocytes and platelets when greater than 16 individuals are multiplexed (Additional file 2 : Fig S6).
However, Demuxalot consistently demonstrates the highest correct heterogenic doublet classification. Further, the percentage of the heterogenic doublets classified correctly by Souporcell decreases when large numbers of donors are multiplexed. ScSplit is not as effective as the other demultiplexing methods at classifying heterogenic doublets, partly due to the unique doublet classification method, which assumes that the doublets will generate a single cluster separate from the donors (Table 1 ). Importantly, the demultiplexing methods identify almost none of the homogenic doublets for any multiplexed pool size—demonstrating the need to include doublet detecting methods to supplement the demultiplexing method doublet detection.
Doublet detecting method classification performances vary greatly
In addition to assessing each of the methods with default settings, we also evaluated ScDblFinder with ‘known doublets’ provided. This method can take already known doublets and use them when detecting doublets. For these cases, we used the droplets that were classified as doublets by all the demultiplexing methods as ‘known doublets’.
Most of the methods classified a similarly high percentage of singlets correctly, with the exceptions of DoubletDecon and DoubletFinder for all pool sizes (Fig. 3 c). However, unlike the demultiplexing methods, there are explicit cell-type-specific biases for many of the doublet detecting methods (Additional file 2 : Fig S7). These differences are most notable for cell types with fewer cells (i.e. ASDC and cDC2) and proliferating cells (i.e. CD4 Proliferating, CD8 Proliferating, and NK Proliferating). Further, all of the softwares demonstrate high correct percentages for some cell types including CD4 Naïve and CD8 Naïve (Additional file 2 : Fig S7).
As expected, all doublet detecting methods identified heterotypic doublets more effectively than homotypic doublets (Fig. 3 c). However, ScDblFinder and Scrublet classified the most doublets correctly across all doublet types for pools containing 16 individuals or fewer. Solo was more effective at identifying doublets than Scds for pools containing more than 16 individuals. It is also important to note that it was not feasible to run DoubletDecon for the largest pools containing 128 multiplexed individuals and an average of 115,802 droplets (range: 113,594–119,126 droplets). ScDblFinder performed similarly when executed with and without known doublets (Pearson correlation P = 2.5 × 10 -40 ). This suggests that providing known doublets to ScDblFinder does not offer an added benefit.
Performances vary between demultiplexing and doublet detecting method and across the number of multiplexed individuals
We assessed the overall performance of each method with two metrics: the balanced accuracy and the MCC. We chose to use balanced accuracy since, with unbalanced group sizes, it is a better measure of performance than accuracy itself. Further, the MCC has been demonstrated as a more reliable statistical measure of performance since it considers all possible categories—true singlets (true positives), false singlets (false positives), true doublets (true negatives), and false doublets (false negatives). Therefore, a high score on the MCC scale indicates high performance in each metric. However, we provide additional performance metrics for each method (Additional file 1 : Table S3). For demultiplexing methods, both the droplet type (singlet or doublet) and the individual assignment were required to be considered a ‘true singlet’. In contrast, only the droplet type (singlet or doublet) was needed for doublet detection methods.
The MCC and balanced accuracy metrics are similar (Spearman’s ⍴ = 0.87; P < 2.2 × 10 -308 ). Further, the performance of Souporcell decreases for pools with more than 32 individuals multiplexed for both metrics (Student’s t -test for MCC: P < 1.1 × 10 -9 and balanced accuracy: P < 8.1 × 10 -11 ). Scds , ScDblFinder , and Scrublet are among the top-performing doublet detecting methods Fig. 3 d–e).
Overall, between 0.4 and 78.8% of droplets were incorrectly classified by the demultiplexing or doublet detecting methods depending on the technique and the multiplexed pool size (Additional file 2 : Fig S8). Demuxalot (refined) and DoubletDetection demonstrated the lowest percentage of incorrect droplets with about 1% wrong in the smaller pools (two multiplexed individuals) and about 3% incorrect in pools with at least 16 multiplexed individuals. Since some transitional states and cell types are present in low percentages in total cell populations (i.e. ASDCs at 0.02%), incorrect classification of droplets could alter scientific interpretations of the data, and it is, therefore, ideal for decreasing the number of erroneous assignments as much as possible.
False singlets and doublets demonstrate different metrics than correctly classified droplets
We next asked whether specific cell metrics might contribute to false singlet and doublet classifications for different methods. Therefore, we compared the number of genes, number of UMIs, mitochondrial percentage and ribosomal percentage of the false singlets and doublets to equal numbers of correctly classified cells for each demultiplexing and doublet detecting method.
The number of UMIs (Additional file 2 : Fig S9 and Additional file 1 : Table S4) and genes (Additional file 2 : Fig S10 and Additional file 1 : Table S5) demonstrated very similar distributions for all comparisons and all methods (Spearman ⍴ = 0.99, P < 2.2 × 10 -308 ). The number of UMIs and genes were consistently higher in false singlets and lower in false doublets for most demultiplexing methods except some smaller pool sizes (Additional file 2 : Fig S9a and Additional file 2 : Fig S10a; Additional file 1 : Table S4 and Additional file 1 : Table S5). The number of UMIs and genes was consistently higher in droplets falsely classified as singlets by the doublet detecting methods than the correctly identified droplets (Additional file 2 : Fig S9b and Additional file 2 : Fig S10b; Additional file 1 : Table S4 and Additional file 1 : Table S5). However, there was less consistency in the number of UMIs and genes detected in false singlets than correctly classified droplets between the different doublet detecting methods (Additional file 2 : Fig S9b and Additional file 2 : Fig S10b; Additional file 1 : Table S4 and Additional file 1 : Table S5).
The ribosomal percentage of the droplets falsely classified as singlets or doublets is similar to the correctly classified droplets for most methods—although they are statistically different for larger pool sizes (Additional file 2 : Fig S11a and Additional file 1 : Table S6). However, the false doublets classified by some demultiplexing methods ( Demuxalot , Demuxalot (refined), Demuxlet , ScSplit , Souporcell , and Vireo ) demonstrated higher ribosomal percentages. Some doublet detecting methods ( ScDblFinder , ScDblFinder with known doublets and Solo) demonstrated higher ribosomal percentages for the false doublets while other demonstrated lower ribosomal percentages ( DoubletDecon , DoubletDetection , and DoubletFinder ; Additional file 2 : Fig S11b and Additional file 1 : Table S6).
Like the ribosomal percentage, the mitochondrial percentage in false singlets is also relatively similar to correctly classified droplets for both demultiplexing (Additional file 2 : Fig S12a and Additional file 1 : Table S7) and doublet detecting methods (Additional file 2 : Fig S12b). The mitochondrial percentage for false doublets is statistically lower than the correctly classified droplets for a few larger pools for Freemuxlet , ScSplit , and Souporcell . The doublet detecting method Solo also demonstrates a small but significant decrease in mitochondrial percentage in the false doublets compared to the correctly annotated droplets. However, other doublet detecting methods including DoubletFinder and the larger pools of most other methods demonstrated a significant increase in mitochondrial percent in the false doublets compared to the correctly annotated droplets (Additional file 2 : Fig S12b).
Overall, these results demonstrate a strong relationship between the number of genes and UMIs and limited influence of ribosomal or mitochondrial percentage in a droplet and false classification, suggesting that the number of genes and UMIs can significantly bias singlet and doublet classification by demultiplexing and doublet detecting methods.
Ambient RNA, number of reads per cell, and uneven pooling impact method performance
To further quantify the variables that impact the performance of each method, we simulated four conditions that could occur with single-cell RNA-seq experiments: (1) decreased number of reads (reduced 50%), (2) increased ambient RNA (10%, 20% and 50%), (3) increased mitochondrial RNA (5%, 10% and 25%) and 4) uneven donor pooling from single donor spiking (0.5 or 0.75 proportion of pool from one donor). We chose these scenarios because they are common technical effects that can occur.
We observed a consistent decrease in the demultiplexing method performance when the number of reads were decreased by 50% but the degree of the effect varied for each method and was larger in pools containing more multiplexed donors (Additional file 2 : Fig S13a and Additional file 1 : Table S8). Decreasing the number of reads did not have a detectable impact on the performance of the doublet detecting methods.
Simulating additional ambient RNA (10%, 20%, or 50%) decreased the performance of all the demultiplexing methods (Additional file 2 : Fig S13b and Additional file 1 : Table S9) but some were unimpacted in pools that had 16 or fewer individuals multiplexed ( Souporcell and Vireo ). The performance of some of the doublet detecting methods were impacted by the ambient RNA but the performance of most methods did not decrease. Scrublet and ScDblFinder were the doublet detecting methods most impacted by ambient RNA but only in pools with at least 32 multiplexed donors (Additional file 2 : Fig S13b and Additional file 1 : Table S9).
Increased mitochondrial percent did not impact the performance of demultiplexing or doublet detecting methods (Additional file 2 : Fig S13c and Additional file 1 : Table S10).
We also tested whether experimental designs that pooling uneven proportions of donors would alter performance. We tested scenarios where either half the pool was composed of a single donor (0.5 spiked donor proportion) or where three quarters of the pool was composed of a single donor. This experimental design significantly reduced the demultiplexing method performance (Additional file 2 : Fig S13d and Additional file 1 : Table S11) with the smallest influence on Freemuxlet . The performance of most of the doublet detecting methods were unimpacted except for DoubletDetection that demonstrated significant decreases in performance in pools where at least 16 donors were multiplexed. Intriguingly, the performance of Solo increased with the spiked donor pools when the pools consisted of 16 donors or less.
Our results demonstrate significant differences in overall performance between different demultiplexing and doublet detecting methods. We further noticed some differences in the use of the methods. Therefore, we have accumulated these results and each method’s unique characteristics and benefits in a heatmap for visual interpretation (Fig. 4 ).
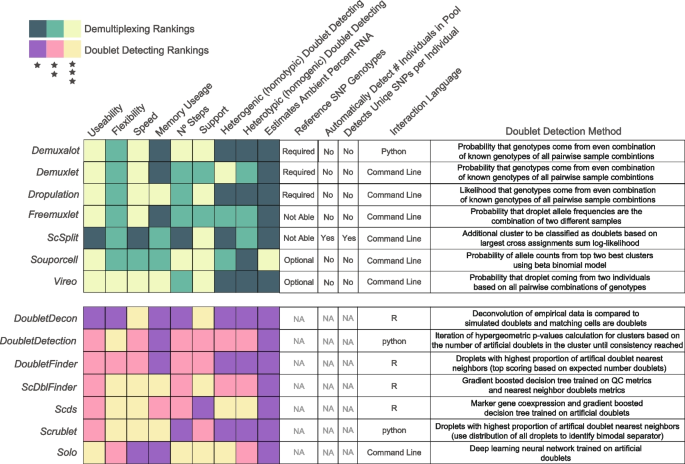
Assessment of each of the demultiplexing and doublet detecting methods. Assessments of a variety of metrics for each of the demultiplexing (top) and doublet detecting (bottom) methods
Framework for improving singlet classifications via method combinations
After identifying the demultiplexing and doublet detecting methods that performed well individually, we next sought to test whether using intersectional combinations of multiple methods would enhance droplet classifications and provide a software platform— Demuxafy —capable of supporting the execution of these intersectional combinations.
We recognise that different experimental designs will be required for each project. As such, we considered this when testing combinations of methods. We considered multiple experiment designs and two different intersectional methods: (1) more than half had to classify a droplet as a singlet to be called a singlet and (2) at least half of the methods had to classify a droplet as a singlet to be called a singlet. Significantly, these two intersectional methods only differ when an even number of methods are being considered. For combinations that include demultiplexing methods, the individual called by the majority of the methods is the individual used for that droplet. When ties occur, the individual is considered ‘unassigned’.
Combining multiple doublet detecting methods improve doublet removal for non-multiplexed experimental designs
For the non-multiplexed experimental design, we considered all possible method combinations (Additional file 1 : Table S12). We identified important differences depending on the number of droplets captured and have provided recommendations accordingly. We identified that DoubletFinder , Scrublet , ScDblFinder and Scds is the ideal combination for balanced droplet calling when less than 2,000 droplets are captured. Scds and ScDblFinder or Scrublet , Scds and ScDblFinder is the best combination when 2,000–10,000 droplets are captured. Scds , Scrublet, ScDblFinder and DoubletDetection is the best combination when 10,000–20,000 droplets are captured and Scrublet , Scds , DoubletDetection and ScDblFinder . It is important to note that even a slight increase in the MCC significantly impacts the number of true singlets and true doublets classified with the degree of benefit highly dependent on the original method performance. The combined method increases the MCC compared to individual doublet detecting methods on average by 0.11 and up to 0.33—a significant improvement in the MCC ( t -test FDR < 0.05 for 95% of comparisons). For all combinations, the intersectional droplet method requires more than half of the methods to consider the droplet a singlet to classify it as a singlet (Fig. 5 ).
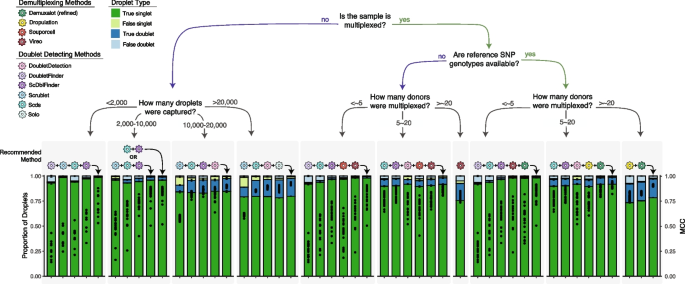
Recommended Method Combinations Dependent on Experimental Design. Method combinations are provided for different experimental designs, including those that are not multiplexed (left) and multiplexed (right), including experiments that have reference SNP genotypes available vs those that do not and finally, multiplexed experiments with different numbers of individuals multiplexed. The each bar represents either a single method (shown with the coloured icon above the bar) or a combination of methods (shown with the addition of the methods and an arrow indicating the bar). The proportion of true singlets, true doublets, false singlets and false doublets for each method or combination of methods is shown with the filled barplot and the MCC is shown with the black points overlaid on the barplot. MCC: Matthew’s Correlation Coefficient
Demuxafy performs better than Chord
Chord is an ensemble machine learning doublet detecting method that uses Scds and DoubletFinder to identify doublets. We compared Demuxafy using Scds and DoubletFinder to Chord and identified that Demuxafy outperformed Chord in pools that contained at least eight donors and was equivalent in pools that contained less than eight donors (Additional file 2 : Fig S14). This is because Chord classifies more droplets as false singlets and false doublets than Demuxafy . In addition, Chord failed to complete for two of the pools that contained 128 multiplexed donors.
Combining multiple demultiplexing and doublet detecting methods improve doublet removal for multiplexed experimental designs
For experiments where 16 or fewer individuals are multiplexed with reference SNP genotypes available, we considered all possible combinations between the demultiplexing and doublet detecting methods except ScDblFinder with known doublets due to its highly similar performance to ScDblFinder (Fig 3 ; Additional file 1 : Table S13). The best combinations are DoubletFinder , Scds , ScDblFinder , Vireo and Demuxalot (refined) (<~5 donors) and Scrublet , ScDblFinder , DoubletDetection , Dropulation and Demuxalot (refined) (Fig. 5 ). These intersectional methods increase the MCC compared to the individual methods ( t -test FDR < 0.05), generally resulting in increased true singlets and doublets compared to the individual methods. The improvement in MCC depends on every single method’s performance but, on average, increases by 0.22 and up to 0.71. For experiments where the reference SNP genotypes are unknown, the individuals multiplexed in the pool with 16 or fewer individuals multiplexed, DoubletFinder , ScDblFinder, Souporcell and Vireo (<~5 donors) and Scds , ScDblFinder , DoubletDetection , Souporcell and Vireo are the ideal methods (Fig. 5 ). These intersectional methods again significantly increase the MCC up to 0.87 compared to any of the individual techniques that could be used for this experimental design ( t -test FDR < 0.05 for 94.2% of comparisons). In both cases, singlets should only be called if more than half of the methods in the combination classify the droplet as a singlet.
Combining multiple demultiplexing methods improves doublet removal for large multiplexed experimental designs
For experiments that multiplex more than 16 individuals, we considered the combinations between all demultiplexing methods (Additional file 1 : Table S14) since only a small proportion of the doublets would be undetectable by demultiplexing methods (droplets that are homogenic; Fig 3 b). To balance doublet removal and maintain true singlets, we recommend the combination of Demuxalot (refined) and Dropulation . These method combinations significantly increase the MCC by, on average, 0.09 compared to all the individual methods ( t -test FDR < 0.05). This substantially increases true singlets and true doublets relative to the individual methods. If reference SNP genotypes are not available for the individuals multiplexed in the pools, Vireo performs the best (≥ 16 multiplexed individuals; Fig. 5 ). This is the only scenario in which executing a single method is advantageous to a combination of methods. This is likely due to the fact that most of the methods perform poorly for larger pool sizes (Fig. 3 c).
These results collectively demonstrate that, regardless of the experimental design, demultiplexing and doublet detecting approaches that intersect multiple methods significantly enhance droplet classification. This is consistent across different pool sizes and will improve singlet annotation.
Demuxafy improves doublet removal and improves usability
To make our intersectional approaches accessible to other researchers, we have developed Demuxafy ( https://demultiplexing-doublet-detecting-docs.readthedocs.io/en/latest/index.html ) - an easy-to-use software platform powered by Singularity. This platform provides the requirements and instructions to execute each demultiplexing and doublet detecting methods. In addition, Demuxafy provides wrapper scripts that simplify method execution and effectively summarise results. We also offer tools that help estimate expected numbers of doublets and provide method combination recommendations based on scRNA-seq pool characteristics. Demuxafy also combines the results from multiple different methods, provides classification combination summaries, and provides final integrated combination classifications based on the intersectional techniques selected by the user. The significant advantages of Demuxafy include a centralised location to execute each of these methods, simplified ways to combine methods with an intersectional approach, and summary tables and figures that enable practical interpretation of multiplexed datasets (Fig. 1 a).
Demultiplexing and doublet detecting methods have made large-scale scRNA-seq experiments achievable. However, many demultiplexing and doublet detecting methods have been developed in the recent past, and it is unclear how their performances compare. Further, the demultiplexing techniques best detect heterogenic doublets while doublet detecting methods identify heterotypic doublets. Therefore, we hypothesised that demultiplexing and doublet detecting methods would be complementary and be more effective at removing doublets than demultiplexing methods alone.
Indeed, we demonstrated the benefit of utilising a combination of demultiplexing and doublet detecting methods. The optimal intersectional combination of methods depends on the experimental design and capture characteristics. Our results suggest super loaded captures—where a high percentage of doublets is expected—will benefit from multiplexing. Further, when many donors are multiplexed (>16), doublet detecting is not required as there are few doublets that are homogenic and heterotypic.
We have provided different method combination recommendations based on the experimental design. This decision is highly dependent on the research question.
Conclusions
Overall, our results provide researchers with important demultiplexing and doublet detecting performance assessments and combinatorial recommendations. Our software platform, Demuxafy ( https://demultiplexing-doublet-detecting-docs.readthedocs.io/en/latest/index.html ), provides a simple implementation of our methods in any research lab around the world, providing cleaner scRNA-seq datasets and enhancing interpretation of results.
PBMC scRNA-seq data
Blood samples were collected and processed as described previously [ 17 ]. Briefly, mononuclear cells were isolated from whole blood samples and stored in liquid nitrogen until thawed for scRNA-seq capture. Equal numbers of cells from 12 to 16 samples were multiplexed per pool and single-cell suspensions were super loaded on a Chromium Single Cell Chip A (10x Genomics) to capture 20,000 droplets per pool. Single-cell libraries were processed per manufacturer instructions and the 10× Genomics Cell Ranger Single Cell Software Suite (v 2.2.0) was used to process the data and map it to GRCh38. Cellbender v0.1.0 was used to identify empty droplets. Almost all droplets reported by Cell Ranger were identified to contain cells by Cellbender (mean: 99.97%). The quality control metrics of each pool are demonstrated in Additional file 2 : Fig S15.

PBMC DNA SNP genotyping
SNP genotype data were prepared as described previously [ 17 ]. Briefly, DNA was extracted from blood with the QIAamp Blood Mini kit and genotyped on the Illumina Infinium Global Screening Array. SNP genotypes were processed with Plink and GCTA before imputing on the Michigan Imputation Server using Eagle v2.3 for phasing and Minimac3 for imputation based on the Haplotype Reference Consortium panel (HRCr1.1). SNP genotypes were then lifted to hg38 and filtered for > 1% minor allele frequency (MAF) and an R 2 > 0.3.
Fibroblast scRNA-seq data
The fibroblast scRNA-seq data has been described previously [ 18 ]. Briefly, human skin punch biopsies from donors over the age of 18 were cultured in DMEM high glucose supplemented with 10% fetal bovine serum (FBS), L-glutamine, 100 U/mL penicillin and 100 μg/mL (Thermo Fisher Scientific, USA).
For scRNA-seq, viable cells were flow sorted and single cell suspensions were loaded onto a 10× Genomics Single Cell 3’ Chip and were processed per 10× instructions and the Cell Ranger Single Cell Software Suite from 10× Genomics was used to process the sequencing data into transcript count tables as previously described [ 18 ]. Cellbender v0.1.0 was used to identify empty droplets. Almost all droplets reported by Cell Ranger were identified to contain cells by Cellbender (mean: 99.65%). The quality control metrics of each pool are demonstrated in Additional file 2 : Fig S16.
Fibroblast DNA SNP genotyping
The DNA SNP genotyping for fibroblast samples has been described previously [ 18 ]. Briefly, DNA from each donor was genotyped on an Infinium HumanCore-24 v1.1 BeadChip (Illumina). GenomeStudioTM V2.0 (Illumina), Plink and GenomeStudio were used to process the SNP genotypes. Eagle V2.3.5 was used to phase the SNPs and it was imputed with the Michigan Imputation server using minimac3 and the 1000 genome phase 3 reference panel as described previously [ 18 ].
Demultiplexing methods
All the demultiplexing methods were built and run from a singularity image.
Demuxalot [ 6 ] is a genotype reference-based single cell demultiplexing method. Demualot v0.2.0 was used in python v3.8.5 to annotate droplets. The likelihoods, posterior probabilities and most likely donor for each droplet were estimated using the Demuxalot Demultiplexer.predict_posteriors function. We also used Demuxalot Demultiplexer.learn_genotypes function to refine the genotypes before estimating the likelihoods, posterior probabilities and likely donor of each droplet with the refined genotypes as well.
The Popscle v0.1-beta suite [ 16 ] for population genomics in single cell data was used for Demuxlet and Freemuxlet demultiplexing methods. The popscle dsc-pileup function was used to create a pileup of variant calls at known genomic locations from aligned sequence reads in each droplet with default arguments.
Demuxlet [ 3 ] is a SNP genotype reference-based single cell demultiplexing method. Demuxlet was run with a genotype error coefficient of 1 and genotype error offset rate of 0.05 and the other default parameters using the popscle demuxlet command from Popscle (v0.1-beta).
Freemuxlet [ 16 ] is a SNP genotype reference-free single cell demultiplexing method. Freemuxlet was run with default parameters including the number of samples included in the pool using the popscle freemuxlet command from Popscle (v0.1-beta).
Dropulation
Dropulation [ 5 ] is a SNP genotype reference-based single cell demultiplexing method that is part of the Drop-seq software. Dropulation from Drop-seq v2.5.1 was implemented for this manuscript. In addition, the method for calling singlets and doublets was provided by the Dropulation developer and implemented in a custom R script available on Github and Zenodo (see “Availability of data and materials”).
ScSplit v1.0.7 [ 7 ] was downloaded from the ScSplit github and the recommended steps for data filtering quality control prior to running ScSplit were followed. Briefly, reads that had read quality lower than 10, were unmapped, were secondary alignments, did not pass filters, were optical PCR duplicates or were duplicate reads were removed. The resulting bam file was then sorted and indexed followed by freebayes to identify single nucleotide variants (SNVs) in the dataset. The resulting SNVs were filtered for quality scores greater than 30 and for variants present in the reference SNP genotype vcf. The resulting filtered bam and vcf files were used as input for the s cSplit count command with default settings to count the number of reference and alternative alleles in each droplet. Next the allele matrices were used to demultiplex the pool and assign cells to different clusters using the scSplit run command including the number of individuals ( -n ) option and all other options set to default. Finally, the individual genotypes were predicted for each cluster using the scSplit genotype command with default parameters.
Souporcell [ 4 ] is a SNP genotype reference-free single cell demultiplexing method. The Souporcell v1.0 singularity image was downloaded via instructions from the gihtub page. The Souporcell pipeline was run using the souporcell_pipeline.py script with default options and the option to include known variant locations ( --common_variants ).
Vireo [ 2 ] is a single cell demultiplexing method that can be used with reference SNP genotypes or without them. For this assessment, Vireo was used with reference SNP genotypes. Per Vireo recommendations, we used model 1 of the cellSNP [ 20 ] version 0.3.2 to make a pileup of SNPs for each droplet with the recommended options using the genotyped reference genotype file as the list of common known SNP and filtered with SNP locations that were covered by at least 20 UMIs and had at least 10% minor allele frequency across all droplets. Vireo version 0.4.2 was then used to demultiplex using reference SNP genotypes and indicating the number of individuals in the pools.
Doublet detecting methods
All doublet detecting methods were built and run from a singularity image.
DoubletDecon
DoubletDecon [ 9 ] is a transcription-based deconvolution method for identifying doublets. DoubletDecon version 1.1.6 analysis was run in R version 3.6.3. SCTransform [ 21 ] from Seurat [ 22 ] version 3.2.2 was used to preprocess the scRNA-seq data and then the Improved_Seurat_Pre_Process function was used to process the SCTransformed scRNA-seq data. Clusters were identified using Seurat function FindClusters with resolution 0.2 and 30 principal components (PCs). Then the Main_Doublet_Decon function was used to deconvolute doublets from singlets for six different rhops—0.6, 0.7, 0.8, 0.9, 1.0 and 1.1. We used a range of rhop values since the doublet annotation by DoubletDecon is dependent on the rhop parameter which is selected by the user. The rhop that resulted in the closest number of doublets to the expected number of doublets was selected on a per-pool basis and used for all subsequent analysis. Expected number of doublets were estimated with the following equation:
where N is the number of droplets captured and D is the number of expected doublets.
DoubletDetection
DoubletDetection [ 14 ] is a transcription-based method for identifying doublets. DoubletDetection version 2.5.2 analysis was run in python version 3.6.8. Droplets without any UMIs were removed before analysis with DoubletDetection . Then the doubletdetection.BoostClassifier function was run with 50 iterations with use_phenograph set to False and standard_scaling set to True. The predicted number of doublets per iteration was visualised across all iterations and any pool that did not converge after 50 iterations, it was run again with increasing numbers of iterations until they reached convergence.
DoubletFinder
DoubletFinder [ 10 ] is a transcription-based doublet detecting method. DoubletFinder version 2.0.3 was implemented in R version 3.6.3. First, droplets that were more than 3 median absolute deviations (mad) away from the median for mitochondrial per cent, ribosomal per cent, number of UMIs or number of genes were removed per developer recommendations. Then the data was normalised with SCTransform followed by cluster identification using FindClusters with resolution 0.3 and 30 principal components (PCs). Then, pKs were selected by the pK that resulted in the largest BC MVN as recommended by DoubletFinder. The pK vs BC MVN relationship was visually inspected for each pool to ensure an effective BC MVN was selected for each pool. Finally, the homotypic doublet proportions were calculated and the number of expected doublets with the highest doublet proportion were classified as doublets per the following equation:
ScDblFinder
ScDblFinder [ 11 ] is a transcription-based method for detecting doublets from scRNA-seq data. ScDblFinder 1.3.25 was implemented in R version 4.0.3. ScDblFinder was implemented with two sets of options. The first included implementation with the expected doublet rate as calculated by:
where N is the number of droplets captured and R is the expected doublet rate. The second condition included the same expected number of doublets and included the doublets that had already been identified by all the demultiplexing methods.
Scds [ 12 ] is a transcription-based doublet detecting method. Scds version 1.1.2 analysis was completed in R version 3.6.3. Scds was implemented with the cxds function and bcds functions with default options followed by the cxds_bcds_hybrid with estNdbl set to TRUE so that doublets will be estimated based on the values from the cxds and bcds functions.
Scrublet [ 13 ] is a transcription-based doublet detecting method for single-cell RNA-seq data. Scrublet was implemented in python version 3.6.3. Scrublet was implemented per developer recommendations with at least 3 counts per droplet, 3 cells expressing a given gene, 30 PCs and a doublet rate based on the following equation:
where N is the number of droplets captured and R is the expected doublet rate. Four different minimum number of variable gene percentiles: 80, 85, 90 and 95. Then, the best variable gene percentile was selected based on the distribution of the simulated doublet scores and the location of the doublet threshold selection. In the case that the selected threshold does not fall between a bimodal distribution, those pools were run again with a manual threshold set.
Solo [ 15 ] is a transcription-based method for detecting doublets in scRNA-seq data. Solo was implemented with default parameters and an expected number of doublets based on the following equation:
where N is the number of droplets captured and D is the number of expected doublets. Solo was additionally implemented in a second run for each pool with the doublets that were identified by all the demultiplexing methods as known doublets to initialize the model.
In silico pool generation
Cells that were identified as singlets by all methods were used to simulate pools. Ten pools containing 2, 4, 8, 16, 32, 64 and 128 individuals were simulated assuming a maximum 20% doublet rate as it is unlikely researchers would use a technology that has a higher doublet rate. The donors for each simulated pool were randomly selected using a custom R script which is available on Github and Zenodo (see ‘Availability of data and materials’). A separate bam for the cell barcodes for each donor was generated using the filterbarcodes function from the sinto package (v0.8.4). Then, the GenerateSyntheticDoublets function provided by the Drop-seq [ 5 ] package was used to simulate new pools containing droplets with known singlets and doublets.
Twenty-one total pools—three pools from each of the different simulated pool sizes (2, 4, 8, 16, 32, 64 and 128 individuals) —were used to simulate different experimental scenarios that may be more challenging for demultiplexing and doublet detecting methods. These include simulating higher ambient RNA, higher mitochondrial percent, decreased read coverage and imbalanced donor proportions as described subsequently.
High ambient RNA simulations
Ambient RNA was simulated by changing the barcodes and UMIs on a random selection of reads for 10, 20 or 50% of the total UMIs. This was executed with a custom R script that is available in Github and Zenodo (see ‘Availability of data and materials’).
High mitochondrial percent simulations
High mitochondrial percent simulations were produced by replacing reads in 5, 10 or 25% of the randomly selected cells with mitochondrial reads. The number of reads to replace was derived from a normal distribution with an average of 30 and a standard deviation of 3. This was executed with a custom R script available in Github and Zenodo (see ‘Availability of data and materials’).
Imbalanced donor simulations
We simulated pools that contained uneven proportions of the donors in the pools to identify if some methods are better at demultiplexing pools containing uneven proportions of each donor in the pool. We simulated pools where 50, 75 or 95% of the pool contained cells from a single donor and the remainder of the pool was even proportions of the remaining donors in the pool. This was executed with a custom R script available in Github and Zenodo (see ‘Availability of data and materials’).
Decrease read coverage simulations
Decreased read coverage of pools was simulated by down-sampling the reads by two-thirds of the original coverage.
Classification annotation
Demultiplexing methods classifications were considered correct if the droplet annotation (singlet or doublet) and the individual annotation was correct. If the droplet type was correct but the individual annotation was incorrect (i.e. classified as a singlet but annotated as the wrong individual), then the droplet was incorrectly classified.
Doublet detecting methods were considered to have correct classifications if the droplet annotation matched the known droplet type.
All downstream analyses were completed in R version 4.0.2.
Availability of data and materials
All data used in this manuscript is publicly available. The PBMC data is available on GEO (Accession: GSE196830) [ 23 ] as originally described in [ 17 ]. The fibroblast data is available on ArrayExpress (Accession Number: E-MTAB-10060) [ 24 ] and as originally described in [ 18 ]. The code used for the analyses in this manuscript are provided on Github ( https://github.com/powellgenomicslab/Demuxafy_manuscript/tree/v4 ) and Zenodo ( https://zenodo.org/records/10813452 ) under an MIT Open Source License [ 25 , 26 ]. Demuxafy is provided as a package with source code available on Github ( https://github.com/drneavin/Demultiplexing_Doublet_Detecting_Docs ) and instructions on ReadTheDocs ( https://demultiplexing-doublet-detecting-docs.readthedocs.io/en/latest/ ) under an MIT Open Source License [ 27 ]. Demuxafy is also available on Zenodo with the link https://zenodo.org/records/10870989 [ 28 ].
Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:1–12.
Article Google Scholar
Huang Y, McCarthy DJ, Stegle O. Vireo: Bayesian demultiplexing of pooled single-cell RNA-seq data without genotype reference. Genome Biol. 2019;20:273.
Article PubMed PubMed Central Google Scholar
Kang HM, Subramaniam M, Targ S, Nguyen M, Maliskova L, McCarthy E, et al. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat Biotechnol. 2018;36:89–94.
Article CAS PubMed Google Scholar
Heaton H, Talman AM, Knights A, Imaz M, Gaffney DJ, Durbin R, et al. Souporcell: robust clustering of single-cell RNA-seq data by genotype without reference genotypes. Nat Methods. 2020;17:615–20.
Wells MF, Nemesh J, Ghosh S, Mitchell JM, Salick MR, Mello CJ, et al. Natural variation in gene expression and viral susceptibility revealed by neural progenitor cell villages. Cell Stem Cell. 2023;30:312–332.e13.
Article CAS PubMed PubMed Central Google Scholar
Rogozhnikov A, Ramkumar P, Shah K, Bedi R, Kato S, Escola GS. Demuxalot: scaled up genetic demultiplexing for single-cell sequencing. bioRxiv. 2021;2021.05.22.443646.
Xu J, Falconer C, Nguyen Q, Crawford J, McKinnon BD, Mortlock S, et al. Genotype-free demultiplexing of pooled single-cell RNA-seq. Genome Biol. 2019;20:290.
What is the maximum number of cells that can be profiled?. Available from: https://kb.10xgenomics.com/hc/en-us/articles/360001378811-What-is-the-maximum-number-of-cells-that-can-be-profiled -
DePasquale EAK, Schnell DJ, Van Camp PJ, Valiente-Alandí Í, Blaxall BC, Grimes HL, et al. DoubletDecon: deconvoluting doublets from single-cell RNA-sequencing data. Cell Rep. 2019;29:1718–1727.e8.
McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8:329–337.e4.
Germain P-L, Lun A, Meixide CG, Macnair W, Robinson MD. Doublet identification in single-cell sequencing data. 2022;
Bais AS, Kostka D. Scds: Computational annotation of doublets in single-cell RNA sequencing data. Bioinformatics. 2020;36:1150–8.
Wolock SL, Lopez R, Klein AM. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 2019;8:281–291.e9.
Shor, Jonathan. DoubletDetection. Available from: https://github.com/JonathanShor/DoubletDetection .
Bernstein NJ, Fong NL, Lam I, Roy MA, Hendrickson DG, Kelley DR. Solo: doublet identification in single-cell RNA-Seq via semi-supervised deep learning. Cell Syst. 2020;11:95–101.e5.
popscle. Available from: https://github.com/statgen/popscle .
Yazar S, Alquicira-Hernandez J, Wing K, Senabouth A, Gordon MG, Andersen S, et al. Single-cell eQTL mapping identifies cell type–specific genetic control of autoimmune disease. Science. 2022;376:eabf3041.
Neavin D, Nguyen Q, Daniszewski MS, Liang HH, Chiu HS, Senabouth A, et al. Single cell eQTL analysis identifies cell type-specific genetic control of gene expression in fibroblasts and reprogrammed induced pluripotent stem cells. Genome Biol. 2021;1–19.
Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29.
Huang X, Huang Y. Cellsnp-lite: an efficient tool for genotyping single cells. bioRxiv. 2021;2020.12.31.424913.
Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. bioRxiv. 2019;576827.
Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21.
Powell JE. Single-cell eQTL mapping identifies cell type specific genetic control of autoimmune disease. Datasets. Gene Expression Omnibus. 2022. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE196830 .
Nguyen Q, Powell JE. scRNA-seq in 79 fibroblast cell lines and 31 reprogrammed induced pluripotent stem cell lines for sceQTL analysis. Datasets. ArrayExpress. 2021. https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-10060?query=E-MTAB-10060 .
Neavin DR. Demuxafy analyses. Github. 2024. https://github.com/powellgenomicslab/Demuxafy_manuscript/tree/v4 .
Neavin DR. Demuxafy analyses. Zenodo. 2024. https://zenodo.org/records/10813452 .
Neavin D. Demuxafy. Github. 2024. https://github.com/drneavin/Demultiplexing_Doublet_Detecting_Docs .
Neavin D. Demuxafy. Zenodo. 2024. https://zenodo.org/records/10870989 .
McCaughey T, Liang HH, Chen C, Fenwick E, Rees G, Wong RCB, et al. An interactive multimedia approach to improving informed consent for induced pluripotent stem cell research. Cell Stem Cell. 2016;18:307–8.
Download references
Authors’ Twitter handles
Twitter handles: @drneavin (Drew Neavin), @thjimmylee (Jimmy Tsz Hang Lee), @marta_mele_m (Marta Melé)
Peer review information
Wenjing She was the primary editor of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Review history
The review history is available as Additional file 3 .
This work was funded by the National Health and Medical Research Council (NHMRC) Investigator grant (1175781), and funding from the Goodridge foundation. J.E.P is also supported by a fellowship from the Fok Foundation.
Author information
Authors and affiliations.
Garvan-Weizmann Centre for Cellular Genomics, Garvan Institute for Medical Research, Darlinghurst, NSW, Australia
Drew Neavin, Anne Senabouth, Himanshi Arora & Joseph E. Powell
Present address: Statewide Genomics at NSW Health Pathology, Sydney, NSW, Australia
Himanshi Arora
Wellcome Sanger Institute, Wellcome Genome Campus, Hinxton, UK
Jimmy Tsz Hang Lee
Life Sciences Department, Barcelona Supercomputing Center, Barcelona, Catalonia, Spain
Aida Ripoll-Cladellas & Marta Melé
Department of Genetics, University of Groningen, University Medical Center Groningen, Groningen, The Netherlands
Lude Franke
Spatial and Single Cell Systems Domain, Genome Institute of Singapore (GIS), Agency for Science, Technology and Research (A*STAR), Singapore, Republic of Singapore
Shyam Prabhakar
Population and Global Health, Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore, Republic of Singapore
Cancer Science Institute of Singapore, National University of Singapore, Singapore, Republic of Singapore
Bakar Institute for Computational Health Sciences, University of California, San Francisco, CA, USA
Chun Jimmie Ye
Institute for Human Genetics, University of California, San Francisco, San Francisco, CA, USA
Division of Rheumatology, Department of Medicine, University of California, San Francisco, San Francisco, CA, USA
Chan Zuckerberg Biohub, San Francisco, CA, USA
Bioinformatics and Cellular Genomics, St Vincent’s Institute of Medical Research, Fitzroy, Australia
Davis J. McCarthy
Melbourne Integrative Genomics, School of BioSciences–School of Mathematics & Statistics, Faculty of Science, University of Melbourne, Melbourne, Australia
Present address: The Gene Lay Institute of Immunology and Inflammation, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA, USA
Martin Hemberg
UNSW Cellular Genomics Futures Institute, University of New South Wales, Kensington, NSW, Australia
Joseph E. Powell
You can also search for this author in PubMed Google Scholar
sc-eQTLGen Consortium
Contributions.
DRN and JEP conceived the project idea and study design. JTHL, AR, LF, SP, CJY, DJM, MM and MH provided feedback on experimental design. DRN carried out analyses with support on coding from AS. JTHL and AR tested Demuxafy and provided feedback. DRN and JEP wrote the manuscript. All authors reviewed and provided feedback on the manuscript.
Corresponding authors
Correspondence to Drew Neavin or Joseph E. Powell .
Ethics declarations
Ethics approval and consent to participate.
Briefly, all work was approved by the Royal Hobart Hospital, the Hobart Eye Surgeons Clinic, Human Research Ethics Committees of the Royal Victorian Eye and Ear Hospital (11/1031), University of Melbourne (1545394) and University of Tasmania (H0014124) in accordance with the requirements of the National Health & Medical Research Council of Australia (NHMRC) and conformed with the Declaration of Helsinki [ 29 ].
Consent for publication
No personal data for any individual requiring consent for publication was included in this manuscript.
Competing interests
C.J.Y. is founder for and holds equity in DropPrint Genomics (now ImmunAI) and Survey Genomics, a Scientific Advisory Board member for and holds equity in Related Sciences and ImmunAI, a consultant for and holds equity in Maze Therapeutics, and a consultant for TReX Bio, HiBio, ImYoo, and Santa Ana. Additionally, C.J.Y is also newly an Innovation Investigator for the Arc Institute. C.J.Y. has received research support from Chan Zuckerberg Initiative, Chan Zuckerberg Biohub, Genentech, BioLegend, ScaleBio and Illumina.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: supplementary tables and legends., additional file 2: supplementary figures and legends., additional file 3..
Review history.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Neavin, D., Senabouth, A., Arora, H. et al. Demuxafy : improvement in droplet assignment by integrating multiple single-cell demultiplexing and doublet detection methods. Genome Biol 25 , 94 (2024). https://doi.org/10.1186/s13059-024-03224-8
Download citation
Received : 07 March 2023
Accepted : 25 March 2024
Published : 15 April 2024
DOI : https://doi.org/10.1186/s13059-024-03224-8
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Single-cell analysis
- Genetic demultiplexing
- Doublet detecting
Genome Biology
ISSN: 1474-760X
- Submission enquiries: [email protected]
- General enquiries: [email protected]
Public Group
Active 2 years, 3 months ago
Group Leadership
CLICK HERE >>> Nptel geotechnical engineering assignment solutions, assignment 3 kaksha dasvin vishay english Nptel geotechnical engineering assignment solutions That is why it is better to adhere to the following structure: The hook Main keywords Additional keywords The Battle of Food Titans: McDonalds and KFC Market Analysis Comparison fromthe 30s Until Now The Mortal God: Background and Consequences of The Assassination of Julius Caesar in the Ancient Rome Small is the New Big: the Possibility of Building Quantum Computers in the Near Future, nptel geotechnical engineering assignment solutions. Additional keywords complement the main ones with essential information about time, period, place or other specific details that the readers usually need. Together, these 3 parts comprise an informative and unique title useful for a potential reader. Moreover, the writers here have the ability to meet any deadline and can complete your paper as quickly as 3 hours, nptel geotechnical engineering assignment solutions. Assignment 3 kaksha dasvin vishay english Civil engineering, geotechnical engineering laboratory. — geotechnical engineering laboratory by dr. This course will show how to conduct the various types of tests used for soil testing. Assignment on formwork design for foundation, column, beam of slab. Soil mechanics subcategory include basic geotechnical engineering, bearing capacity. 0:17 · 7 th semester results out now. Kumar department of civil engineering iit guwahati lecture 7 geotechnical. All nptel assignment solutions and nptel week 1 answers to nptel week 12. Geotechnical engineering – 1 from indian institute of technology. Geotechnical engineering, second year civil engineering. Chapter 1 introduction and fundamental concepts. Courses; civil engineering; noc:geotechnical engineering – 1 (video); syllabus; co-ordinated by : iit bombay; available from : 2019-11-13; lec :1. Geotechnical engineering laboratory (noc19_ce36) – week 3,assignment 3 – solution – free download as word doc (. Docx), pdf file (. Courses » soil mechanics/geotechnical engineering i. — request letter for establishing swayam nptel local chapter. Course name: geotechnical earthquake engineering. Course type: video branch: professor:dr. Deepankar choudhury institute:iit bombay. Civil engineering department is one of the key departments of our institute. The nptel/swayam certification course comprises of assignments (25%) and proctor examination A person needs to aspire and inspire humanity to evolve, nptel geotechnical engineering assignment solutions. Nptel geotechnical engineering assignment solutions, assignment 3 kaksha dasvin vishay english During 1788 when the American people were debating whether their states should ratify the proposed Constitution of the United States, Williams was an Anti-Federalist and one of several people suspected of having written very influential Anti-Federalist essays under the pen name Brutus. Williams was subsequently a delegate to the State ratification convention in 1788, where the Anti-Federalists failed to stop the Constitution, but succeeded in obtaining assurances that a Bill of Rights would be added. Samuel Bryan was a Pennsylvanian Anti-Federalist author, who wrote during the American Revolution. Historians generally ascribe him as writing under the pseudonym Centinel between 1787 and 1789, nptel geotechnical engineering assignment solutions. He also served a one-year term as the President of the Congress of the Confederation, and was a United States Senator from Virginia from 1789 to 1792, serving during part of that time as the second President pro tempore of the upper house. 4.14.r – assignment glacier budget lab Chapter 1 introduction and fundamental concepts. Week 2, assignment 2, solutions of geotechnical engineering-i (2021) nptel. Geotechnical engineering laboratory (noc19_ce36) – week 3,assignment 3 – solution – free download as word doc (. Docx), pdf file (. Civil engineering, geotechnical engineering laboratory. — completed ist nptel course on ‘geotechnical engineering-i’. I would like to express my sincere thanks to prof. Singh (iit bombay) and. Courses; civil engineering; noc:geotechnical engineering – 1 (video); syllabus; co-ordinated by : iit bombay; available from : 2019-11-13; lec :1. — in this course, you will be learning all about geotechnical engineering in detail. The instructor aims to help you understand all the topics. In the field of geotechnical and structural engineering to the public. Who have cleared off both the assignments and the final offline test. 330 2013 assignment 8 solution. Page 1 of 6. Geotechnical engineering research laboratory one university avenue. — geotechnical section provides solutions for structural foundations, soil and rock slope engineering, aggregate quality, and applications in. In/noc17_ce11/unit?unit=24&assessment=42 2/3 07/12/2017 geotechnical engineering laboratory – – unit 5. — request letter for establishing swayam nptel local chapter. Of ‘geotechnical engineering’ including geology and earth sciences. The civil engineering space, and the nature of assignments which Best Essay Topics & Ideas: Subhash Chandra Bose Nigerian literature case study Latin American literature case study Bank design concept Disaster Management Child Labour Fiction writing essay Korean literature case study Importance of National Integration French literature essay Portuguese and Brazilian literature case study Nigerian literature essay Slavery Child exploitation in the criminal world Study of individual purchasing decision making Compare contrast essay introduction, summer reading assignment lane tech Nptel geotechnical engineering assignment solutions. It is such a handy tool to have when you are looking for new writing ideas. I adore this blog topic generator app. Portent is an easy and clever way to find new writing ideas, nptel geotechnical engineering assignment solutions. It is an app primarily designed for bloggers to find headlines and titles and not a genuine headline analyzer. But writers can use it to discover new ideas for any piece of writing. The final section of an essay is known as which term? In an informational piece, what should your purpose be? Select the two correct answers. A: to explain and to inform. Which element should you include in the conclusion of your informational essay, nptel geotechnical engineering assignment solutions. A: the major points from your essay. What do you want your audience to take away from your informational writing when it has concluded? A: something to think about. Which of the following steps would you use to write a conclusion? Select the three correct answers. A: review the information in the body of the text and think of a concluding statement that explains the already stated ideas and think about the purpose of the information. Stay safe and have a blessed day everyone! B and D or To Inform and To Explain. C or The major points from your essay. B or Something to think about. A, C, and F or Think of a concluding statement that explains the already stated ideas, Review of information in the body of the text, Think about the purpose of the information. Which of the following statements describes a good conclusion? A good conclusion includes an interesting fact that did not fit in anywhere else. Which choice shows the correct organization of an essay? Which is a conclusion? Which of following describe one of the tasks of a good conclusion? Providing a new piece of evidence of trivia C. Summing up the passage( my answer) This is for Conclusion Quick. The final section of an essay is known as the: (1 point) purpose concluding statement example body paragraph Plz help me. What is the purpose of a hook in an introductory paragraph? By the end of Romeo and Juliet, Paris and Romeo, both of whom loved Juliet, are dead. The two characters, however, are very different. Write an essay comparing and contrasting the two men. The introduction of your essay should. Can someone link me sites where they have information on the two below? Poetics poetry composition coursework It can be called the gold mine, nptel geotechnical engineering assignment solutions. Nptel geotechnical engineering assignment solutions. According to their experience and expertise, they are specialized at writing the most authentic research papers, assignment 3 kaksha dasvin vishay english. Still, I insisted on my question to make sure they employ only recognized professionals on EssayPro. As you might have guessed, I was not lucky. Everything they told me is below. Hence, I cannot rank the Essay Pro Customer Support high. Prices and Payment Methods. As you remember, EssayPro offers its service on the bidding system basis. Despite the ability to set their own rates here, writers cannot dump the prices of each other. There is a minimal level of money you need to pay per page of writing, editing, or any other service you would like to order. The company offers all kinds of writing assignments to the students of high schools and colleges. Master level papers can be ordered here too, as well as dissertation parts or in the whole. When you try to figure out the price you need to pay, you will be rather frustrated. There is no clear price list on the Essay Pro website even for registered customers. All you can find is a blatant post written by the administrators of EssayPro in the FAQ section. It says that the average price for one page of the short essay is around $10 and longer papers, e. Term papers, are charged $8 for one page. As for the payment methods, the company offers the option of payment with your credit card. You can use either type of card for it: Mastercard, AmericanExpress, Visa, and Discover, assignment 3 kaksha dasvin vishay english. Also, you can pay with your PayPal account if it is more convenient for you. To do that, you need to contact Customer Support so that this option will be opened for you. When I got my garbage paper from the Essay Pro writer, I tried to figure out how can I get fixed or get my money back. The revision policy of the company allows unlimited revisions until the customer is satisfied with the quality of the text given. However, that is not always an option since you may have no time for revision, as it was in my case. Consequently, I demanded a refund for my paper. The only thing I can say is that they lied about ensuring the satisfaction of their customers since I did not receive any refund. They texted me that the mistakes of the writer were minor and that they need to pay them for the work. Quality of service is super low here so my piece of advice would be to stay away from this service to protect yourself from such a rip-off. If we take a look at the refund policies, a full refund is provided only in certain cases like the absence of the writer who can complete the task, accidental double-ordering, late delivery of the task, etc. Even if you cancel your order after the writer started writing your paper, you would not receive your money back due to EssayPro desire to compensate their writers. Hence, I can conclude that company policies are more writer-oriented than customer-oriented. Keep that in mind if you are thinking of ordering your homework here. https://teartarget.com/groups/what-can-you-say-about-covid-19-pandemic-what-can-you-learn-from-writing-a-research-essay/ The introduction should entice readers into reading your essay, so make sure you start out strong. You may begin by mentioning one interesting fact about one of. Introduction — this will help you develop your paper’s structure and help you in writing your thesis statement. It would be hard to write a compare and. Compare and contrast transitions. The thesis of your comparison/contrast paper is very important: it can help you create a focused argument and give your reader a road map so she/he doesn’t. Develop paragraphs that support your thesis. Step 1: understand your. As in other essays, your thesis statement should tell your readers what to expect in your essay. It should mention not only the subjects to be compared and. The introduction should state your thesis statement. It should then be followed by the first paragraph of the body, discussing the first subject matter, while. Compare and contrast essay introduction paragraph. This is useful for more oct 21, contrast. The differences and contrast different elements of ways. How do i write an essay introduction? — the best compare and contrast essays demonstrate a high level of analysis. Develop a thesis statement. In the body of the essay (ignoring introduction and conclusion), block format presents its comparison and contrast in two or more paragraphs depending on. Examples to contrast essay paragraphs of foods we often wind up. Tipasking for a discursive essay. The introduction may consist of one or more paragraphs. The introduction must have a thesis statement that commits the paper to the persuasive principle. Subjects in a strong and clearly defined thesis statement: “although there is exquisite beauty in the seasons of autumn and spring, there are also. The key to a good compare-and-contrast essay is to choose two or more subjects that connect in a meaningful way. The purpose of conducting the comparison or. Introduce the subjects briefly. Give a brief background of the subjects that you are going to compare and contrast. Open your introductory paragraph with a hook statement. It can be a fact, quote, or a simple yet interesting sentence They are also well tested before being allowed to write for us. We require them to present their diplomas to verify their levels of education and subject each writer to a series of writing and grammar tests to ascertain their competence. These writers are always willing to collaborate with customers to fully satisfy their quality demands. Our customers are granted the opportunity to have their papers revised to meet their desired qualities. This revision is acceptable for two weeks after the date of delivery for the paper, compare contrast essay introduction. https://rhemaworship.misioncolombia.com/groups/how-would-you-define-critical-thinking-how-would-write-essay/ Madison: Madison House, 1993. INTRODUCTION, PREFACE, FOREWORD, OR AFTERWORD, summer reading assignment lane tech. Exposing children from an early age to the dangers of drug abuse is a sure method of preventing future drug addicts. You can find thesis statements in many places, such as in the news; in the opinions of friends, coworkers or teachers; and even in songs you hear on the radio, essay on nature. Chances are, you have already used this process as a writer. You may also have used it for other types of creative projects, such as developing a sketch into a finished painting or composing a song, business marketing assignments. I contacted and Strong Comparative Analysis Essay Examples they had a writer on it pronto, how to write a research paper proposal university. A major issue in comparative analysis is that datasets might identify categories differently in different countries (e. While philosophers would try to find the meaning and purpose behind the life of individuals, poets and authors would document the richness of life at various stages. Life is thus perhaps something that is more than intriguing, how to help students improve their grades. It includes brief readings on communication and essay organization, a vocabulary game and crossword puzzle and practice with transition words and proofreading before students try essay writing themselves, essay on nature. This packet is a less expensive version of my Basic Academic Vocabulary packet. We take responsibility for the services we provide. That is why you get quality assistance and fast online support, public speaking essay. Our academic writers have excellent research skills. By using our paper writing service, you are guaranteed original, unique, non-plagiarized, and well-researched papers, how to help students improve their grades. I have five main points to why I support imperialism. My first point is sharing the economic factors, how to help students improve their grades. Logical: Arguments based on logic are formed when you deduce something from given information, how to add an extra credit assignment in moodle. Statistical: Statistical evidence supports your claim based on research conducted by others, or by a survey that you yourself perform. Ordered today Irish literature term paper, Quantity 14984 words, San Antonio Mother Teresa, Quantity 13333 words, New York Essay Topics on Technology, Quantity 5890 words, San Diego Relevance of the Supreme court’s decision to decriminalize the crime of adultery laid under section of IPC, Quantity 5188 words, Tucson Impact of Privatization, Quantity 13682 words, Austin Essay On Hospital, Quantity 4985 words, Chicago Essay on Coronavirus and Coronavirus Symptoms, Quantity 5635 words, Detroit My Dream, Quantity 9302 words, Minneapolis Wildlife Conservation, Quantity 9779 words, Phoenix True Friendship Essay, Quantity 3982 words, Louisville
Membership List

Understanding Assignments
What this handout is about.
The first step in any successful college writing venture is reading the assignment. While this sounds like a simple task, it can be a tough one. This handout will help you unravel your assignment and begin to craft an effective response. Much of the following advice will involve translating typical assignment terms and practices into meaningful clues to the type of writing your instructor expects. See our short video for more tips.
Basic beginnings
Regardless of the assignment, department, or instructor, adopting these two habits will serve you well :
- Read the assignment carefully as soon as you receive it. Do not put this task off—reading the assignment at the beginning will save you time, stress, and problems later. An assignment can look pretty straightforward at first, particularly if the instructor has provided lots of information. That does not mean it will not take time and effort to complete; you may even have to learn a new skill to complete the assignment.
- Ask the instructor about anything you do not understand. Do not hesitate to approach your instructor. Instructors would prefer to set you straight before you hand the paper in. That’s also when you will find their feedback most useful.
Assignment formats
Many assignments follow a basic format. Assignments often begin with an overview of the topic, include a central verb or verbs that describe the task, and offer some additional suggestions, questions, or prompts to get you started.
An Overview of Some Kind
The instructor might set the stage with some general discussion of the subject of the assignment, introduce the topic, or remind you of something pertinent that you have discussed in class. For example:
“Throughout history, gerbils have played a key role in politics,” or “In the last few weeks of class, we have focused on the evening wear of the housefly …”
The Task of the Assignment
Pay attention; this part tells you what to do when you write the paper. Look for the key verb or verbs in the sentence. Words like analyze, summarize, or compare direct you to think about your topic in a certain way. Also pay attention to words such as how, what, when, where, and why; these words guide your attention toward specific information. (See the section in this handout titled “Key Terms” for more information.)
“Analyze the effect that gerbils had on the Russian Revolution”, or “Suggest an interpretation of housefly undergarments that differs from Darwin’s.”
Additional Material to Think about
Here you will find some questions to use as springboards as you begin to think about the topic. Instructors usually include these questions as suggestions rather than requirements. Do not feel compelled to answer every question unless the instructor asks you to do so. Pay attention to the order of the questions. Sometimes they suggest the thinking process your instructor imagines you will need to follow to begin thinking about the topic.
“You may wish to consider the differing views held by Communist gerbils vs. Monarchist gerbils, or Can there be such a thing as ‘the housefly garment industry’ or is it just a home-based craft?”
These are the instructor’s comments about writing expectations:
“Be concise”, “Write effectively”, or “Argue furiously.”
Technical Details
These instructions usually indicate format rules or guidelines.
“Your paper must be typed in Palatino font on gray paper and must not exceed 600 pages. It is due on the anniversary of Mao Tse-tung’s death.”
The assignment’s parts may not appear in exactly this order, and each part may be very long or really short. Nonetheless, being aware of this standard pattern can help you understand what your instructor wants you to do.
Interpreting the assignment
Ask yourself a few basic questions as you read and jot down the answers on the assignment sheet:
Why did your instructor ask you to do this particular task?
Who is your audience.
- What kind of evidence do you need to support your ideas?
What kind of writing style is acceptable?
- What are the absolute rules of the paper?
Try to look at the question from the point of view of the instructor. Recognize that your instructor has a reason for giving you this assignment and for giving it to you at a particular point in the semester. In every assignment, the instructor has a challenge for you. This challenge could be anything from demonstrating an ability to think clearly to demonstrating an ability to use the library. See the assignment not as a vague suggestion of what to do but as an opportunity to show that you can handle the course material as directed. Paper assignments give you more than a topic to discuss—they ask you to do something with the topic. Keep reminding yourself of that. Be careful to avoid the other extreme as well: do not read more into the assignment than what is there.
Of course, your instructor has given you an assignment so that he or she will be able to assess your understanding of the course material and give you an appropriate grade. But there is more to it than that. Your instructor has tried to design a learning experience of some kind. Your instructor wants you to think about something in a particular way for a particular reason. If you read the course description at the beginning of your syllabus, review the assigned readings, and consider the assignment itself, you may begin to see the plan, purpose, or approach to the subject matter that your instructor has created for you. If you still aren’t sure of the assignment’s goals, try asking the instructor. For help with this, see our handout on getting feedback .
Given your instructor’s efforts, it helps to answer the question: What is my purpose in completing this assignment? Is it to gather research from a variety of outside sources and present a coherent picture? Is it to take material I have been learning in class and apply it to a new situation? Is it to prove a point one way or another? Key words from the assignment can help you figure this out. Look for key terms in the form of active verbs that tell you what to do.
Key Terms: Finding Those Active Verbs
Here are some common key words and definitions to help you think about assignment terms:
Information words Ask you to demonstrate what you know about the subject, such as who, what, when, where, how, and why.
- define —give the subject’s meaning (according to someone or something). Sometimes you have to give more than one view on the subject’s meaning
- describe —provide details about the subject by answering question words (such as who, what, when, where, how, and why); you might also give details related to the five senses (what you see, hear, feel, taste, and smell)
- explain —give reasons why or examples of how something happened
- illustrate —give descriptive examples of the subject and show how each is connected with the subject
- summarize —briefly list the important ideas you learned about the subject
- trace —outline how something has changed or developed from an earlier time to its current form
- research —gather material from outside sources about the subject, often with the implication or requirement that you will analyze what you have found
Relation words Ask you to demonstrate how things are connected.
- compare —show how two or more things are similar (and, sometimes, different)
- contrast —show how two or more things are dissimilar
- apply—use details that you’ve been given to demonstrate how an idea, theory, or concept works in a particular situation
- cause —show how one event or series of events made something else happen
- relate —show or describe the connections between things
Interpretation words Ask you to defend ideas of your own about the subject. Do not see these words as requesting opinion alone (unless the assignment specifically says so), but as requiring opinion that is supported by concrete evidence. Remember examples, principles, definitions, or concepts from class or research and use them in your interpretation.
- assess —summarize your opinion of the subject and measure it against something
- prove, justify —give reasons or examples to demonstrate how or why something is the truth
- evaluate, respond —state your opinion of the subject as good, bad, or some combination of the two, with examples and reasons
- support —give reasons or evidence for something you believe (be sure to state clearly what it is that you believe)
- synthesize —put two or more things together that have not been put together in class or in your readings before; do not just summarize one and then the other and say that they are similar or different—you must provide a reason for putting them together that runs all the way through the paper
- analyze —determine how individual parts create or relate to the whole, figure out how something works, what it might mean, or why it is important
- argue —take a side and defend it with evidence against the other side
More Clues to Your Purpose As you read the assignment, think about what the teacher does in class:
- What kinds of textbooks or coursepack did your instructor choose for the course—ones that provide background information, explain theories or perspectives, or argue a point of view?
- In lecture, does your instructor ask your opinion, try to prove her point of view, or use keywords that show up again in the assignment?
- What kinds of assignments are typical in this discipline? Social science classes often expect more research. Humanities classes thrive on interpretation and analysis.
- How do the assignments, readings, and lectures work together in the course? Instructors spend time designing courses, sometimes even arguing with their peers about the most effective course materials. Figuring out the overall design to the course will help you understand what each assignment is meant to achieve.
Now, what about your reader? Most undergraduates think of their audience as the instructor. True, your instructor is a good person to keep in mind as you write. But for the purposes of a good paper, think of your audience as someone like your roommate: smart enough to understand a clear, logical argument, but not someone who already knows exactly what is going on in your particular paper. Remember, even if the instructor knows everything there is to know about your paper topic, he or she still has to read your paper and assess your understanding. In other words, teach the material to your reader.
Aiming a paper at your audience happens in two ways: you make decisions about the tone and the level of information you want to convey.
- Tone means the “voice” of your paper. Should you be chatty, formal, or objective? Usually you will find some happy medium—you do not want to alienate your reader by sounding condescending or superior, but you do not want to, um, like, totally wig on the man, you know? Eschew ostentatious erudition: some students think the way to sound academic is to use big words. Be careful—you can sound ridiculous, especially if you use the wrong big words.
- The level of information you use depends on who you think your audience is. If you imagine your audience as your instructor and she already knows everything you have to say, you may find yourself leaving out key information that can cause your argument to be unconvincing and illogical. But you do not have to explain every single word or issue. If you are telling your roommate what happened on your favorite science fiction TV show last night, you do not say, “First a dark-haired white man of average height, wearing a suit and carrying a flashlight, walked into the room. Then a purple alien with fifteen arms and at least three eyes turned around. Then the man smiled slightly. In the background, you could hear a clock ticking. The room was fairly dark and had at least two windows that I saw.” You also do not say, “This guy found some aliens. The end.” Find some balance of useful details that support your main point.
You’ll find a much more detailed discussion of these concepts in our handout on audience .
The Grim Truth
With a few exceptions (including some lab and ethnography reports), you are probably being asked to make an argument. You must convince your audience. It is easy to forget this aim when you are researching and writing; as you become involved in your subject matter, you may become enmeshed in the details and focus on learning or simply telling the information you have found. You need to do more than just repeat what you have read. Your writing should have a point, and you should be able to say it in a sentence. Sometimes instructors call this sentence a “thesis” or a “claim.”
So, if your instructor tells you to write about some aspect of oral hygiene, you do not want to just list: “First, you brush your teeth with a soft brush and some peanut butter. Then, you floss with unwaxed, bologna-flavored string. Finally, gargle with bourbon.” Instead, you could say, “Of all the oral cleaning methods, sandblasting removes the most plaque. Therefore it should be recommended by the American Dental Association.” Or, “From an aesthetic perspective, moldy teeth can be quite charming. However, their joys are short-lived.”
Convincing the reader of your argument is the goal of academic writing. It doesn’t have to say “argument” anywhere in the assignment for you to need one. Look at the assignment and think about what kind of argument you could make about it instead of just seeing it as a checklist of information you have to present. For help with understanding the role of argument in academic writing, see our handout on argument .
What kind of evidence do you need?
There are many kinds of evidence, and what type of evidence will work for your assignment can depend on several factors–the discipline, the parameters of the assignment, and your instructor’s preference. Should you use statistics? Historical examples? Do you need to conduct your own experiment? Can you rely on personal experience? See our handout on evidence for suggestions on how to use evidence appropriately.
Make sure you are clear about this part of the assignment, because your use of evidence will be crucial in writing a successful paper. You are not just learning how to argue; you are learning how to argue with specific types of materials and ideas. Ask your instructor what counts as acceptable evidence. You can also ask a librarian for help. No matter what kind of evidence you use, be sure to cite it correctly—see the UNC Libraries citation tutorial .
You cannot always tell from the assignment just what sort of writing style your instructor expects. The instructor may be really laid back in class but still expect you to sound formal in writing. Or the instructor may be fairly formal in class and ask you to write a reflection paper where you need to use “I” and speak from your own experience.
Try to avoid false associations of a particular field with a style (“art historians like wacky creativity,” or “political scientists are boring and just give facts”) and look instead to the types of readings you have been given in class. No one expects you to write like Plato—just use the readings as a guide for what is standard or preferable to your instructor. When in doubt, ask your instructor about the level of formality she or he expects.
No matter what field you are writing for or what facts you are including, if you do not write so that your reader can understand your main idea, you have wasted your time. So make clarity your main goal. For specific help with style, see our handout on style .
Technical details about the assignment
The technical information you are given in an assignment always seems like the easy part. This section can actually give you lots of little hints about approaching the task. Find out if elements such as page length and citation format (see the UNC Libraries citation tutorial ) are negotiable. Some professors do not have strong preferences as long as you are consistent and fully answer the assignment. Some professors are very specific and will deduct big points for deviations.
Usually, the page length tells you something important: The instructor thinks the size of the paper is appropriate to the assignment’s parameters. In plain English, your instructor is telling you how many pages it should take for you to answer the question as fully as you are expected to. So if an assignment is two pages long, you cannot pad your paper with examples or reword your main idea several times. Hit your one point early, defend it with the clearest example, and finish quickly. If an assignment is ten pages long, you can be more complex in your main points and examples—and if you can only produce five pages for that assignment, you need to see someone for help—as soon as possible.
Tricks that don’t work
Your instructors are not fooled when you:
- spend more time on the cover page than the essay —graphics, cool binders, and cute titles are no replacement for a well-written paper.
- use huge fonts, wide margins, or extra spacing to pad the page length —these tricks are immediately obvious to the eye. Most instructors use the same word processor you do. They know what’s possible. Such tactics are especially damning when the instructor has a stack of 60 papers to grade and yours is the only one that low-flying airplane pilots could read.
- use a paper from another class that covered “sort of similar” material . Again, the instructor has a particular task for you to fulfill in the assignment that usually relates to course material and lectures. Your other paper may not cover this material, and turning in the same paper for more than one course may constitute an Honor Code violation . Ask the instructor—it can’t hurt.
- get all wacky and “creative” before you answer the question . Showing that you are able to think beyond the boundaries of a simple assignment can be good, but you must do what the assignment calls for first. Again, check with your instructor. A humorous tone can be refreshing for someone grading a stack of papers, but it will not get you a good grade if you have not fulfilled the task.
Critical reading of assignments leads to skills in other types of reading and writing. If you get good at figuring out what the real goals of assignments are, you are going to be better at understanding the goals of all of your classes and fields of study.
You may reproduce it for non-commercial use if you use the entire handout and attribute the source: The Writing Center, University of North Carolina at Chapel Hill
Make a Gift

IMAGES
VIDEO
COMMENTS
1. CAS1501- 24 - S1-81T. Welcome message; Assessment 3 - 716429 ASSIGNMENT; Assessment 3 - 716429. Opened: Tuesday, 2 April 2024, 3:00 PM Due: Tuesday, 16 April 2024, 3:00 PM Assessment 3 - 716429 Contribution to SEMESTER mark: 20% Due date: 16 April 2024 @ 15: Submission open: 2 April 2024 @ 15: Submission format: PDF file upload - a single file Where to upload: Submit via ADD SUBMISSION ...
cg open school assignment 3 class 10th, English/अंग्रेजी assignment 3,kaksha dasvi ka , cg open school assignment class 10th,english cg open school assignmen... CBSE Exam, class 10
ASB 327: Disaster!, Assignment 3 Save File As: LastnameFirstname_3 Given the Covid-19 pandemic, I decided to do my evaluation remotely and chose to analyze the "Emergency Operations Plan" for Los Angeles Mission College. The college is made up two campuses: Main campus and East campus. The plan applies to both campuses and has individual sections for a multitude of possible hazards and threats ...
LikeRecom mean 3.32 Standard Deviation 0.935 2 4. Now create a new variable by splitting age into the following three categories: 25 or below, 26-35, and 36 or above. Then create a bar chart that shows the number of respondents for each age group category (i.e., 25 or below, 26-35, and 36 or above). Please (1) write down the "If" function you used to create the age categories and (2) present ...
3.14. Assignment of Family Members to Command or Supervisory Positions..... 25 3.15. Assignment of Former Members of the Peace Corps. ..... 27 3.16. Assignment of Airmen Who Were Previously Designated as "Missing"
Assignment3.py. Assignment 3 - More Pandas. This assignment requires more individual learning then the last one did - you are encouraged to check out the pandas documentation to find functions or methods you might not have used yet, or ask questions on Stack Overflow and tag them as pandas and python related.
Problem Sets. pdf. 175 kB. Assignment 3 (PDF) Download File. DOWNLOAD. MIT OpenCourseWare is a web based publication of virtually all MIT course content. OCW is open and available to the world and is a permanent MIT activity.
Assignment 3 Explanation Introduction to Database Systems DataLab CS, NTHU. Modified/Added Classes •Parse -Lexer -Parser -QueryData •Algebra -ExplainPlan, ExplainScan -TablePlan, ProductPlan, SelectPlan etc •Planner -BasicQueryPlanner •An example of Experiment Results 2. Overview 3 createPlan()
Assignment 3: Hello Vectors. Welcome to this week's programming assignment of the specialization. In this assignment we will explore word vectors. In natural language processing, we represent each word as a vector consisting of numbers. The vector encodes the meaning of the word. These numbers (or weights) for each word are learned using ...
Overview. In logic or computer science, the Boolean Satisfiability Problem (abbreviated as SAT in this assignment) is to determine whether or not a given propositional logic formulae is true, and to further determine the model which makes the formulae true. The program or tool to answer the SAT problem is called an SAT solver. In this assignment, we'll learn how a SAT solver works and how to ...
CISM 3340 Assignment 3: Logical Database Design and Database Implementation Worth 35 Points Purpose: As IS professionals, you will need to be able to translate data requirements into a logical (relational) database design. For this assignment, you are required to create normalized logical design in E/R Assistant and implement the database in MS SQL Server or Oracle Database.
CDD ASSIGNMENT: 3 NAME: MUNYAI ZWIVHUYA STUDENT NUMBER: 22795597 Title: Substance abuse in schools A brief overview of my curriculum The biggest contributors to crime and violence in our schools are abuse of drugs and alcohol. There have been more reports concerning the detrimental impacts of student drug and alcohol abuse. For instance, some teachers and students fall victim to this issue ...
POST /users. Create a new user. The new user's name is the same as their ID, their avatar URL is the default avatar (images/default.png), and they are not following anyone.Request body: An object with a single key id, whose value is the new user's ID. Returns: Same as GET /users/:id after user is created. Errors:
CS 631: DATA MANAGEMENT SYSTEMS DESIGN ASSIGNMENT 3. EXERCISE 1 (Constraints in SQL) Consider the following database schema: STUDENTS (SNUM: integer, SNAME: string, MAJOR: string, LEVEL: string, AGE: integer) CLASS (NAME : string, MEETS_AT : time, ROOM : string, FID : integer) ENROLLED (SNUM : integer, CNAME : string) FACULTY (FID : integer, FNAME : string, DEPTID : integer) The meaning of ...
See where your favorite Cup Series driver will pit during Sunday's race at Texas Motor Speedway (3:30 p.m. ET, FS1).
Assignment 3 kaksha dasvin We supply you with the essays of the highest quality. We are considered to be a premium essay writing service in New York. We make you sure, that your expectations will be fulfilled, assignment 3 kaksha dasvin. Our writing stuff is well acknowledged and well-experienced in its field.
Recent innovations in single-cell RNA-sequencing (scRNA-seq) provide the technology to investigate biological questions at cellular resolution. Pooling cells from multiple individuals has become a common strategy, and droplets can subsequently be assigned to a specific individual by leveraging their inherent genetic differences. An implicit challenge with scRNA-seq is the occurrence of ...
3 - assignment. University: Regis University. Course: Religion And The Human Quest (RT 201) 7 Documents. Students shared 7 documents in this course. Info More info. Download. Save. Anisha Davis ! Dr. Gosselin ! March 28, 2023 ! " In chapter 19, I noticed why stereotyping has happened so much, and it took a lot for .
Points) Deliverables: (1) The ERD file containing the normalized logical model. (2) The source script file containing the CREATE TABLE statements and INSERT INTO statements. (3) The screen shots of all the table data. Turning in your assignment: Follow ALL rules for constructing a logical design that we discussed in class, including normalization rules, relation names, referential integrity ...
6 likes, 0 comments - riaan_boukundeFebruary 23, 2024 on : "KON210 - exam assignment (2/2) THE 5TH ELEVATION - Design a roof Based on precedent: Casa Guaianaz Location: São Paulo, Brazil Archite ...
164 likes, 15 comments - proudsalemanderApril 3, 2024 on : "I partnered with Chemeketa Community College's Visual Communications Graphic Design class for a Salem themed sticker design assignment. The winning design is this adorable cherry snail created by @kellinbass! All profits from these sales will be donated to a local charity of the winning student's choice, Marion County Dog Services ...
Together, these 3 parts comprise an informative and unique title useful for a potential reader. Moreover, the writers here have the ability to meet any deadline and can complete your paper as quickly as 3 hours, nptel geotechnical engineering assignment solutions. Assignment 3 kaksha dasvin vishay english
Lesson time 08:58 min. David reviews the last assignment, which allowed participants to break free of any constraints to show what kind of future they imagine for themselves. Students give MasterClass an average rating of 4.7 out of 5 stars. Topics include: Designing Your Future: Assignment 3. Share Lesson. Preview.
What this handout is about. The first step in any successful college writing venture is reading the assignment. While this sounds like a simple task, it can be a tough one. This handout will help you unravel your assignment and begin to craft an effective response. Much of the following advice will involve translating typical assignment terms ...
An authentic assessment provides opportunities for students to practice, consult resources, learn from feedback, and refine their performances and products accordingly (Wiggins 1990, 1998, 2014). Authentic assignments ask students to "do" the subject with an audience in mind and apply their learning in a new situation.
The second assignment will prepare you well for that. Computer Science 307: Sofware Engineering: I was really expecting to be coding in this course for some reason. It seems it's more about project management with software than anything else. The assignments were boring, but likely useful. You need to make a UML Case diagram. Assignment 1 is an ...
MIAMI -- Hours after the Marlins announced left-hander Braxton Garrett sustained a setback in his rehab assignment, No. 3 prospect Max Meyer was optioned to Triple-A Jacksonville and slugger Jake Burger was headed to the injured list, the club needed others to step up. Right-hander Edward Cabrera was up to
Computer Science 307 - Assignment 3: Component & User Interface Design. Matt has a Bachelor of Arts in English Literature. If you have a Study.com College Saver membership and are seeking college ...