The top list of academic search engines

1. Google Scholar
4. science.gov, 5. semantic scholar, 6. baidu scholar, get the most out of academic search engines, frequently asked questions about academic search engines, related articles.
Academic search engines have become the number one resource to turn to in order to find research papers and other scholarly sources. While classic academic databases like Web of Science and Scopus are locked behind paywalls, Google Scholar and others can be accessed free of charge. In order to help you get your research done fast, we have compiled the top list of free academic search engines.
Google Scholar is the clear number one when it comes to academic search engines. It's the power of Google searches applied to research papers and patents. It not only lets you find research papers for all academic disciplines for free but also often provides links to full-text PDF files.
- Coverage: approx. 200 million articles
- Abstracts: only a snippet of the abstract is available
- Related articles: ✔
- References: ✔
- Cited by: ✔
- Links to full text: ✔
- Export formats: APA, MLA, Chicago, Harvard, Vancouver, RIS, BibTeX
BASE is hosted at Bielefeld University in Germany. That is also where its name stems from (Bielefeld Academic Search Engine).
- Coverage: approx. 136 million articles (contains duplicates)
- Abstracts: ✔
- Related articles: ✘
- References: ✘
- Cited by: ✘
- Export formats: RIS, BibTeX

CORE is an academic search engine dedicated to open-access research papers. For each search result, a link to the full-text PDF or full-text web page is provided.
- Coverage: approx. 136 million articles
- Links to full text: ✔ (all articles in CORE are open access)
- Export formats: BibTeX
Science.gov is a fantastic resource as it bundles and offers free access to search results from more than 15 U.S. federal agencies. There is no need anymore to query all those resources separately!
- Coverage: approx. 200 million articles and reports
- Links to full text: ✔ (available for some databases)
- Export formats: APA, MLA, RIS, BibTeX (available for some databases)
Semantic Scholar is the new kid on the block. Its mission is to provide more relevant and impactful search results using AI-powered algorithms that find hidden connections and links between research topics.
- Coverage: approx. 40 million articles
- Export formats: APA, MLA, Chicago, BibTeX

Although Baidu Scholar's interface is in Chinese, its index contains research papers in English as well as Chinese.
- Coverage: no detailed statistics available, approx. 100 million articles
- Abstracts: only snippets of the abstract are available
- Export formats: APA, MLA, RIS, BibTeX

RefSeek searches more than one billion documents from academic and organizational websites. Its clean interface makes it especially easy to use for students and new researchers.
- Coverage: no detailed statistics available, approx. 1 billion documents
- Abstracts: only snippets of the article are available
- Export formats: not available
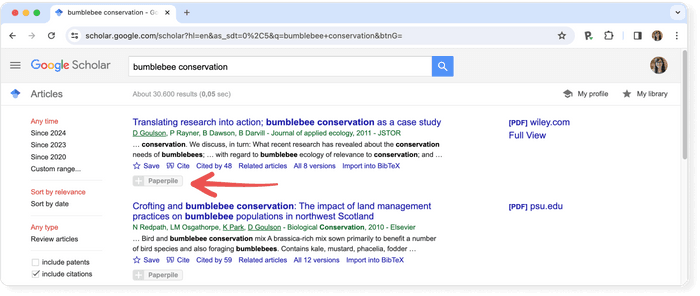
Consider using a reference manager like Paperpile to save, organize, and cite your references. Paperpile integrates with Google Scholar and many popular databases, so you can save references and PDFs directly to your library using the Paperpile buttons:
Google Scholar is an academic search engine, and it is the clear number one when it comes to academic search engines. It's the power of Google searches applied to research papers and patents. It not only let's you find research papers for all academic disciplines for free, but also often provides links to full text PDF file.
Semantic Scholar is a free, AI-powered research tool for scientific literature developed at the Allen Institute for AI. Sematic Scholar was publicly released in 2015 and uses advances in natural language processing to provide summaries for scholarly papers.
BASE , as its name suggest is an academic search engine. It is hosted at Bielefeld University in Germany and that's where it name stems from (Bielefeld Academic Search Engine).
CORE is an academic search engine dedicated to open access research papers. For each search result a link to the full text PDF or full text web page is provided.
Science.gov is a fantastic resource as it bundles and offers free access to search results from more than 15 U.S. federal agencies. There is no need any more to query all those resources separately!
Help | Advanced Search
arXiv is a free distribution service and an open-access archive for nearly 2.4 million scholarly articles in the fields of physics, mathematics, computer science, quantitative biology, quantitative finance, statistics, electrical engineering and systems science, and economics. Materials on this site are not peer-reviewed by arXiv.
arXiv is a free distribution service and an open-access archive for scholarly articles in the fields of physics, mathematics, computer science, quantitative biology, quantitative finance, statistics, electrical engineering and systems science, and economics. Materials on this site are not peer-reviewed by arXiv.
Stay up to date with what is happening at arXiv on our blog.
Latest news
- Astrophysics ( astro-ph new , recent , search ) Astrophysics of Galaxies ; Cosmology and Nongalactic Astrophysics ; Earth and Planetary Astrophysics ; High Energy Astrophysical Phenomena ; Instrumentation and Methods for Astrophysics ; Solar and Stellar Astrophysics
- Condensed Matter ( cond-mat new , recent , search ) Disordered Systems and Neural Networks ; Materials Science ; Mesoscale and Nanoscale Physics ; Other Condensed Matter ; Quantum Gases ; Soft Condensed Matter ; Statistical Mechanics ; Strongly Correlated Electrons ; Superconductivity
- General Relativity and Quantum Cosmology ( gr-qc new , recent , search )
- High Energy Physics - Experiment ( hep-ex new , recent , search )
- High Energy Physics - Lattice ( hep-lat new , recent , search )
- High Energy Physics - Phenomenology ( hep-ph new , recent , search )
- High Energy Physics - Theory ( hep-th new , recent , search )
- Mathematical Physics ( math-ph new , recent , search )
- Nonlinear Sciences ( nlin new , recent , search ) includes: Adaptation and Self-Organizing Systems ; Cellular Automata and Lattice Gases ; Chaotic Dynamics ; Exactly Solvable and Integrable Systems ; Pattern Formation and Solitons
- Nuclear Experiment ( nucl-ex new , recent , search )
- Nuclear Theory ( nucl-th new , recent , search )
- Physics ( physics new , recent , search ) includes: Accelerator Physics ; Applied Physics ; Atmospheric and Oceanic Physics ; Atomic and Molecular Clusters ; Atomic Physics ; Biological Physics ; Chemical Physics ; Classical Physics ; Computational Physics ; Data Analysis, Statistics and Probability ; Fluid Dynamics ; General Physics ; Geophysics ; History and Philosophy of Physics ; Instrumentation and Detectors ; Medical Physics ; Optics ; Physics and Society ; Physics Education ; Plasma Physics ; Popular Physics ; Space Physics
- Quantum Physics ( quant-ph new , recent , search )
Mathematics
- Mathematics ( math new , recent , search ) includes: (see detailed description ): Algebraic Geometry ; Algebraic Topology ; Analysis of PDEs ; Category Theory ; Classical Analysis and ODEs ; Combinatorics ; Commutative Algebra ; Complex Variables ; Differential Geometry ; Dynamical Systems ; Functional Analysis ; General Mathematics ; General Topology ; Geometric Topology ; Group Theory ; History and Overview ; Information Theory ; K-Theory and Homology ; Logic ; Mathematical Physics ; Metric Geometry ; Number Theory ; Numerical Analysis ; Operator Algebras ; Optimization and Control ; Probability ; Quantum Algebra ; Representation Theory ; Rings and Algebras ; Spectral Theory ; Statistics Theory ; Symplectic Geometry
Computer Science
- Computing Research Repository ( CoRR new , recent , search ) includes: (see detailed description ): Artificial Intelligence ; Computation and Language ; Computational Complexity ; Computational Engineering, Finance, and Science ; Computational Geometry ; Computer Science and Game Theory ; Computer Vision and Pattern Recognition ; Computers and Society ; Cryptography and Security ; Data Structures and Algorithms ; Databases ; Digital Libraries ; Discrete Mathematics ; Distributed, Parallel, and Cluster Computing ; Emerging Technologies ; Formal Languages and Automata Theory ; General Literature ; Graphics ; Hardware Architecture ; Human-Computer Interaction ; Information Retrieval ; Information Theory ; Logic in Computer Science ; Machine Learning ; Mathematical Software ; Multiagent Systems ; Multimedia ; Networking and Internet Architecture ; Neural and Evolutionary Computing ; Numerical Analysis ; Operating Systems ; Other Computer Science ; Performance ; Programming Languages ; Robotics ; Social and Information Networks ; Software Engineering ; Sound ; Symbolic Computation ; Systems and Control
Quantitative Biology
- Quantitative Biology ( q-bio new , recent , search ) includes: (see detailed description ): Biomolecules ; Cell Behavior ; Genomics ; Molecular Networks ; Neurons and Cognition ; Other Quantitative Biology ; Populations and Evolution ; Quantitative Methods ; Subcellular Processes ; Tissues and Organs
Quantitative Finance
- Quantitative Finance ( q-fin new , recent , search ) includes: (see detailed description ): Computational Finance ; Economics ; General Finance ; Mathematical Finance ; Portfolio Management ; Pricing of Securities ; Risk Management ; Statistical Finance ; Trading and Market Microstructure
- Statistics ( stat new , recent , search ) includes: (see detailed description ): Applications ; Computation ; Machine Learning ; Methodology ; Other Statistics ; Statistics Theory
Electrical Engineering and Systems Science
- Electrical Engineering and Systems Science ( eess new , recent , search ) includes: (see detailed description ): Audio and Speech Processing ; Image and Video Processing ; Signal Processing ; Systems and Control
- Economics ( econ new , recent , search ) includes: (see detailed description ): Econometrics ; General Economics ; Theoretical Economics
About arXiv
- General information
- How to Submit to arXiv
- Membership & Giving
- Advanced search
- Peer review

Discover relevant research today

Advance your research field in the open

Reach new audiences and maximize your readership
ScienceOpen puts your research in the context of
Publications
For Publishers
ScienceOpen offers content hosting, context building and marketing services for publishers. See our tailored offerings
- For academic publishers to promote journals and interdisciplinary collections
- For open access journals to host journal content in an interactive environment
- For university library publishing to develop new open access paradigms for their scholars
- For scholarly societies to promote content with interactive features
For Institutions
ScienceOpen offers state-of-the-art technology and a range of solutions and services
- For faculties and research groups to promote and share your work
- For research institutes to build up your own branding for OA publications
- For funders to develop new open access publishing paradigms
- For university libraries to create an independent OA publishing environment
For Researchers
Make an impact and build your research profile in the open with ScienceOpen
- Search and discover relevant research in over 94 million Open Access articles and article records
- Share your expertise and get credit by publicly reviewing any article
- Publish your poster or preprint and track usage and impact with article- and author-level metrics
- Create a topical Collection to advance your research field
Create a Journal powered by ScienceOpen
Launching a new open access journal or an open access press? ScienceOpen now provides full end-to-end open access publishing solutions – embedded within our smart interactive discovery environment. A modular approach allows open access publishers to pick and choose among a range of services and design the platform that fits their goals and budget.
Continue reading “Create a Journal powered by ScienceOpen”
What can a Researcher do on ScienceOpen?
ScienceOpen provides researchers with a wide range of tools to support their research – all for free. Here is a short checklist to make sure you are getting the most of the technological infrastructure and content that we have to offer. What can a researcher do on ScienceOpen? Continue reading “What can a Researcher do on ScienceOpen?”
ScienceOpen on the Road
Upcoming events.
- 15 June – Scheduled Server Maintenance, 13:00 – 01:00 CEST
Past Events
- 20 – 22 February – ResearcherToReader Conference
- 09 November – Webinar for the Discoverability of African Research
- 26 – 27 October – Attending the Workshop on Open Citations and Open Scholarly Metadata
- 18 – 22 October – ScienceOpen at Frankfurt Book Fair.
- 27 – 29 September – Attending OA Tage, Berlin .
- 25 – 27 September – ScienceOpen at Open Science Fair
- 19 – 21 September – OASPA 2023 Annual Conference .
- 22 – 24 May – ScienceOpen sponsoring Pint of Science, Berlin.
- 16-17 May – ScienceOpen at 3rd AEUP Conference.
- 20 – 21 April – ScienceOpen attending Scaling Small: Community-Owned Futures for Open Access Books .
What is ScienceOpen?
- Smart search and discovery within an interactive interface
- Researcher promotion and ORCID integration
- Open evaluation with article reviews and Collections
- Business model based on providing services to publishers
Live Twitter stream
Some of our partners:.

A free, AI-powered research tool for scientific literature
- Dennis O'Dea
- Electromagnetism
New & Improved API for Developers
Introducing semantic reader in beta.
Stay Connected With Semantic Scholar Sign Up What Is Semantic Scholar? Semantic Scholar is a free, AI-powered research tool for scientific literature, based at the Allen Institute for AI.
SpringerOpen
The SpringerOpen portfolio has grown tremendously since its launch in 2010, so that we now offer researchers from all areas of science, technology, medicine, the humanities and social sciences a place to publish open access in journals. Publishing with SpringerOpen makes your work freely available online for everyone, immediately upon publication, and our high-level peer-review and production processes guarantee the quality and reliability of the work. Open access books are published by our Springer imprint.
Find the right journal for you
Explore our subject areas, learn all about open access.

- Browse our alphabetical journal list
- Explore our journals by subject
- Tips for finding the right journal
- Find the right journal with our Journal Suggester
- Find out if open access book publishing is right for you

- Visit our subject pages covering all subject areas in science, technology, medicine, the humanities and social sciences

Visit the Springer Author & Reviewer tutorials and learn all about open access, your benefits, mandates, funding, copyright and more in the different interactive tutorials. And take our quiz to test your knowledge!
- C heck out the free tutorials
- Take the quiz
Video library
Your browser needs to have JavaScript enabled to view this video
Our video library contains how-to videos, videos on the research we publish and journal videos.
“The only truly modern academic research engine”
Oa.mg is a search engine for academic papers, specialising in open access. we have over 250 million papers in our index..
21 Legit Research Databases for Free Journal Articles in 2024
#scribendiinc
Written by Scribendi
Has this ever happened to you? While looking for websites for research, you come across a research paper site that claims to connect academics to a peer-reviewed article database for free.
Intrigued, you search for keywords related to your topic, only to discover that you must pay a hefty subscription fee to access the service. After the umpteenth time being duped, you begin to wonder if there's even such a thing as free journal articles.
Subscription fees and paywalls are often the bane of students and academics, especially those at small institutions who don't provide access to many free article directories and repositories.
Whether you're working on an undergraduate paper, a PhD dissertation, or a medical research study, we want to help you find tools to locate and access the information you need to produce well-researched, compelling, and innovative work.
Below, we discuss why peer-reviewed articles are superior and list out the best free article databases to use in 2024.
Download Our Free Research Database Roundup PDF
Why peer-reviewed scholarly journal articles are more authoritative.

Determining what sources are reliable can be challenging. Peer-reviewed scholarly journal articles are the gold standard in academic research. Reputable academic journals have a rigorous peer-review process.
The peer review process provides accountability to the academic community, as well as to the content of the article. The peer review process involves qualified experts in a specific (often very specific) field performing a review of an article's methods and findings to determine things like quality and credibility.
Peer-reviewed articles can be found in peer-reviewed article databases and research databases, and if you know that a database of journals is reliable, that can offer reassurances about the reliability of a free article. Peer review is often double blind, meaning that the author removes all identifying information and, likewise, does not know the identity of the reviewers. This helps reviewers maintain objectivity and impartiality so as to judge an article based on its merit.
Where to Find Peer-Reviewed Articles
Peer-reviewed articles can be found in a variety of research databases. Below is a list of some of the major databases you can use to find peer-reviewed articles and other sources in disciplines spanning the humanities, sciences, and social sciences.
What Are Open Access Journals?
An open access (OA) journal is a journal whose content can be accessed without payment. This provides scholars, students, and researchers with free journal articles. OA journals use alternate methods of funding to cover publication costs so that articles can be published without having to pass those publication costs on to the reader.

Some of these funding models include standard funding methods like advertising, public funding, and author payment models, where the author pays a fee in order to publish in the journal. There are OA journals that have non-peer-reviewed academic content, as well as journals that focus on dissertations, theses, and papers from conferences, but the main focus of OA is peer-reviewed scholarly journal articles.
The internet has certainly made it easier to access research articles and other scholarly publications without needing access to a university library, and OA takes another step in that direction by removing financial barriers to academic content.
Choosing Wisely
Features of legitimate oa journals.
There are things to look out for when trying to decide if a free publication journal is legitimate:
Mission statement —The mission statement for an OA journal should be available on their website.
Publication history —Is the journal well established? How long has it been available?
Editorial board —Who are the members of the editorial board, and what are their credentials?
Indexing —Can the journal be found in a reliable database?
Peer review —What is the peer review process? Does the journal allow enough time in the process for a reliable assessment of quality?
Impact factor —What is the average number of times the journal is cited over a two-year period?
Features of Illegitimate OA Journals
There are predatory publications that take advantage of the OA format, and they are something to be wary of. Here are some things to look out for:
Contact information —Is contact information provided? Can it be verified?
Turnaround —If the journal makes dubious claims about the amount of time from submission to publication, it is likely unreliable.
Editorial board —Much like determining legitimacy, looking at the editorial board and their credentials can help determine illegitimacy.
Indexing —Can the journal be found in any scholarly databases?
Peer review —Is there a statement about the peer review process? Does it fit what you know about peer review?
How to Find Scholarly Articles
Identify keywords.
Keywords are included in an article by the author. Keywords are an excellent way to find content relevant to your research topic or area of interest. In academic searches, much like you would on a search engine, you can use keywords to navigate through what is available to find exactly what you're looking for.
Authors provide keywords that will help you easily find their article when researching a related topic, often including general terms to accommodate broader searches, as well as some more specific terms for those with a narrower scope. Keywords can be used individually or in combination to refine your scholarly article search.
Narrow Down Results
Sometimes, search results can be overwhelming, and searching for free articles on a journal database is no exception, but there are multiple ways to narrow down your results. A good place to start is discipline.
What category does your topic fall into (psychology, architecture, machine learning, etc.)? You can also narrow down your search with a year range if you're looking for articles that are more recent.
A Boolean search can be incredibly helpful. This entails including terms like AND between two keywords in your search if you need both keywords to be in your results (or, if you are looking to exclude certain keywords, to exclude these words from the results).
Consider Different Avenues
If you're not having luck using keywords in your search for free articles, you may still be able to find what you're looking for by changing your tactics. Casting a wider net sometimes yields positive results, so it may be helpful to try searching by subject if keywords aren't getting you anywhere.
You can search for a specific publisher to see if they have OA publications in the academic journal database. And, if you know more precisely what you're looking for, you can search for the title of the article or the author's name.
Determining the Credibility of Scholarly Sources
Ensuring that sources are both credible and reliable is crucial to academic research. Use these strategies to help evaluate the usefulness of scholarly sources:
- Peer Review : Look for articles that have undergone a rigorous peer-review process. Peer-reviewed articles are typically vetted by experts in the field, ensuring the accuracy of the research findings.
Tip: To determine whether an article has undergone rigorous peer review, review the journal's editorial policies, which are often available on the journal's website. Look for information about the peer-review process, including the criteria for selecting reviewers, the process for handling conflicts of interest, and any transparency measures in place.
- Publisher Reputation : Consider the reputation of the publisher. Established publishers, such as well-known academic journals, are more likely to adhere to high editorial standards and publishing ethics.
- Author Credentials : Evaluate the credentials and expertise of the authors. Check their affiliations, academic credentials, and past publications to assess their authority in the field.
- Citations and References : Examine the citations and references provided in the article. A well-researched article will cite credible sources to support its arguments and findings. Verify the accuracy of the cited sources and ensure they are from reputable sources.
- Publication Date : Consider the publication date of the article. While older articles may still be relevant, particularly in certain fields, it is best to prioritize recent publications for up-to-date research and findings.
- Journal Impact Factor : Assess the journal's impact factor or other metrics that indicate its influence and reputation within the academic community. Higher impact factor journals are generally considered more prestigious and reliable.
Tip: Journal Citation Reports (JCR), produced by Clarivate Analytics, is a widely used source for impact factor data. You can access JCR through academic libraries or directly from the Clarivate Analytics website if you have a subscription.
- Peer Recommendations : Seek recommendations from peers, mentors, or professors in your field. They can provide valuable insights and guidance on reputable sources and journals within your area of study.
- Cross-Verification : Cross-verify the information presented in the article with other credible sources. Compare findings, methodologies, and conclusions with similar studies to ensure consistency and reliability.
By employing these strategies, researchers can confidently evaluate the credibility and reliability of scholarly sources, ensuring the integrity of their research contributions in an ever-evolving landscape.
The Top 21 Free Online Journal and Research Databases
Navigating OA journals, research article databases, and academic websites trying to find high-quality sources for your research can really make your head spin. What constitutes a reliable database? What is a useful resource for your discipline and research topic? How can you find and access full-text, peer-reviewed articles?
Fortunately, we're here to help. Having covered some of the ins and outs of peer review, OA journals, and how to search for articles, we have compiled a list of the top 21 free online journals and the best research databases. This list of databases is a great resource to help you navigate the wide world of academic research.
These databases provide a variety of free sources, from abstracts and citations to full-text, peer-reviewed OA journals. With databases covering specific areas of research and interdisciplinary databases that provide a variety of material, these are some of our favorite free databases, and they're totally legit!
CORE is a multidisciplinary aggregator of OA research. CORE has the largest collection of OA articles available. It allows users to search more than 219 million OA articles. While most of these link to the full-text article on the original publisher's site, or to a PDF available for download, five million records are hosted directly on CORE.
CORE's mission statement is a simple and straightforward commitment to offering OA articles to anyone, anywhere in the world. They also host communities that are available for researchers to join and an ambassador community to enhance their services globally. In addition to a straightforward keyword search, CORE offers advanced search options to filter results by publication type, year, language, journal, repository, and author.
CORE's user interface is easy to use and navigate. Search results can be sorted based on relevance or recency, and you can search for relevant content directly from the results screen.
Collection : 219,537,133 OA articles
Other Services : Additional services are available from CORE, with extras that are geared toward researchers, repositories, and businesses. There are tools for accessing raw data, including an API that provides direct access to data, datasets that are available for download, and FastSync for syncing data content from the CORE database.
CORE has a recommender plug-in that suggests relevant OA content in the database while conducting a search and a discovery feature that helps you discover OA versions of paywalled articles. Other features include tools for managing content, such as a dashboard for managing repository output and the Repository Edition service to enhance discoverability.
Good Source of Peer-Reviewed Articles : Yes
Advanced Search Options : Language, author, journal, publisher, repository, DOI, year
2. ScienceOpen
Functioning as a research and publishing network, ScienceOpen offers OA to more than 74 million articles in all areas of science. Although you do need to register to view the full text of articles, registration is free. The advanced search function is highly detailed, allowing you to find exactly the research you're looking for.
The Berlin- and Boston-based company was founded in 2013 to "facilitate open and public communications between academics and to allow ideas to be judged on their merit, regardless of where they come from." Search results can be exported for easy integration with reference management systems.
You can also bookmark articles for later research. There are extensive networking options, including your Science Open profile, a forum for interacting with other researchers, the ability to track your usage and citations, and an interactive bibliography. Users have the ability to review articles and provide their knowledge and insight within the community.
Collection : 74,560,631
Other Services : None
Advanced Search Options : Content type, source, author, journal, discipline
3. Directory of Open Access Journals
A multidisciplinary, community-curated directory, the Directory of Open Access Journals (DOAJ) gives researchers access to high-quality peer-reviewed journals. It has archived more than two million articles from 17,193 journals, allowing you to either browse by subject or search by keyword.
The site was launched in 2003 with the aim of increasing the visibility of OA scholarly journals online. Content on the site covers subjects from science, to law, to fine arts, and everything in between. DOAJ has a commitment to "increase the visibility, accessibility, reputation, usage and impact of quality, peer-reviewed, OA scholarly research journals globally, regardless of discipline, geography or language."
Information about the journal is available with each search result. Abstracts are also available in a collapsible format directly from the search screen. The scholarly article website is somewhat simple, but it is easy to navigate. There are 16 principles of transparency and best practices in scholarly publishing that clearly outline DOAJ policies and standards.
Collection : 6,817,242
Advanced Search Options : Subject, journal, year
4. Education Resources Information Center
The Education Resources Information Center (ERIC) of the Institution of Education Sciences allows you to search by topic for material related to the field of education. Links lead to other sites, where you may have to purchase the information, but you can search for full-text articles only. You can also search only peer-reviewed sources.
The service primarily indexes journals, gray literature (such as technical reports, white papers, and government documents), and books. All sources of material on ERIC go through a formal review process prior to being indexed. ERIC's selection policy is available as a PDF on their website.
The ERIC website has an extensive FAQ section to address user questions. This includes categories like general questions, peer review, and ERIC content. There are also tips for advanced searches, as well as general guidance on the best way to search the database. ERIC is an excellent database for content specific to education.
Collection : 1,292,897
Advanced Search Options : Boolean
5. arXiv e-Print Archive
The arXiv e-Print Archive is run by Cornell University Library and curated by volunteer moderators, and it now offers OA to more than one million e-prints.
There are advisory committees for all eight subjects available on the database. With a stated commitment to an "emphasis on openness, collaboration, and scholarship," the arXiv e-Print Archive is an excellent STEM resource.
The interface is not as user-friendly as some of the other databases available, and the website hosts a blog to provide news and updates, but it is otherwise a straightforward math and science resource. There are simple and advanced search options, and, in addition to conducting searches for specific topics and articles, users can browse content by subject. The arXiv e-Print Archive clearly states that they do not peer review the e-prints in the database.
Collection : 1,983,891
Good Source of Peer-Reviewed Articles : No
Advanced Search Options : Subject, date, title, author, abstract, DOI
6. Social Science Research Network
The Social Science Research Network (SSRN) is a collection of papers from the social sciences community. It is a highly interdisciplinary platform used to search for scholarly articles related to 67 social science topics. SSRN has a variety of research networks for the various topics available through the free scholarly database.
The site offers more than 700,000 abstracts and more than 600,000 full-text papers. There is not yet a specific option to search for only full-text articles, but, because most of the papers on the site are free access, it's not often that you encounter a paywall. There is currently no option to search for only peer-reviewed articles.
You must become a member to use the services, but registration is free and enables you to interact with other scholars around the world. SSRN is "passionately committed to increasing inclusion, diversity and equity in scholarly research," and they encourage and discuss the use of inclusive language in scholarship whenever possible.
Collection : 1,058,739 abstracts; 915,452 articles
Advanced Search Options : Term, author, date, network
7. Public Library of Science
Public Library of Science (PLOS) is a big player in the world of OA science. Publishing 12 OA journals, the nonprofit organization is committed to facilitating openness in academic research. According to the site, "all PLOS content is at the highest possible level of OA, meaning that scientific articles are immediately and freely available to anyone, anywhere."
PLOS outlines four fundamental goals that guide the organization: break boundaries, empower researchers, redefine quality, and open science. All PLOS journals are peer-reviewed, and all 12 journals uphold rigorous ethical standards for research, publication, and scientific reporting.
PLOS does not offer advanced search options. Content is organized by topic into research communities that users can browse through, in addition to options to search for both articles and journals. The PLOS website also has resources for peer reviewers, including guidance on becoming a reviewer and on how to best participate in the peer review process.
Collection : 12 journals
Advanced Search Options : None
8. OpenDOAR
OpenDOAR, or the Directory of Open Access Repositories, is a comprehensive resource for finding free OA journals and articles. Using Google Custom Search, OpenDOAR combs through OA repositories around the world and returns relevant research in all disciplines.
The repositories it searches through are assessed and categorized by OpenDOAR staff to ensure they meet quality standards. Inclusion criteria for the database include requirements for OA content, global access, and categorically appropriate content, in addition to various other quality assurance measures. OpenDOAR has metadata, data, content, preservation, and submission policies for repositories, in addition to two OA policy statements regarding minimum and optimum recommendations.
This database allows users to browse and search repositories, which can then be selected, and articles and data can be accessed from the repository directly. As a repository database, much of the content on the site is geared toward the support of repositories and OA standards.
Collection : 5,768 repositories
Other Services : OpenDOAR offers a variety of additional services. Given the nature of the platform, services are primarily aimed at repositories and institutions, and there is a marked focus on OA in general. Sherpa services are OA archiving tools for authors and institutions.
They also offer various resources for OA support and compliance regarding standards and policies. The publication router matches publications and publishers with appropriate repositories.
There are also services and resources from JISC for repositories for cost management, discoverability, research impact, and interoperability, including ORCID consortium membership information. Additionally, a repository self-assessment tool is available for members.
Advanced Search Options : Name, organization name, repository type, software name, content type, subject, country, region
9. Bielefeld Academic Search Engine
The Bielefeld Academic Search Engine (BASE) is operated by the Bielefeld University Library in Germany, and it offers more than 240 million documents from more than 8,000 sources. Sixty percent of its content is OA, and you can filter your search accordingly.
BASE has rigorous inclusion requirements for content providers regarding quality and relevance, and they maintain a list of content providers for the sake of transparency, which can be easily found on their website. BASE has a fairly elegant interface. Search results can be organized by author, title, or date.
From the search results, items can be selected and exported, added to favorites, emailed, and searched in Google Scholar. There are basic and advanced search features, with the advanced search offering numerous options for refining search criteria. There is also a feature on the website that saves recent searches without additional steps from the user.
Collection : 276,019,066 documents; 9,286 content providers
Advanced Search Options : Author, subject, year, content provider, language, document type, access, terms of reuse

10. Digital Library of the Commons Repository
Run by Indiana University, the Digital Library of the Commons (DLC) Repository is a multidisciplinary journal repository that allows users to access thousands of free and OA articles from around the world. You can browse by document type, date, author, title, and more or search for keywords relevant to your topic.
DCL also offers the Comprehensive Bibliography of the Commons, an image database, and a keyword thesaurus for enhanced search parameters. The repository includes books, book chapters, conference papers, journal articles, surveys, theses and dissertations, and working papers. DCL advanced search features drop-down menus of search types with built-in Boolean search options.
Searches can be sorted by relevance, title, date, or submission date in ascending or descending order. Abstracts are included in selected search results, with access to full texts available, and citations can be exported from the same page. Additionally, the image database search includes tips for better search results.
Collection : 10,784
Advanced Search Options : Author, date, title, subject, sector, region, conference
11. CIA World Factbook
The CIA World Factbook is a little different from the other resources on this list in that it is not an online journal directory or repository. It is, however, a useful free online research database for academics in a variety of disciplines.
All the information is free to access, and it provides facts about every country in the world, which are organized by category and include information about history, geography, transportation, and much more. The World Factbook can be searched by country or region, and there is also information about the world's oceans.
This site contains resources related to the CIA as an organization rather than being a scientific journal database specifically. The site has a user interface that is easy to navigate. The site also provides a section for updates regarding changes to what information is available and how it is organized, making it easier to interact with the information you are searching for.
Collection : 266 countries
12. Paperity
Paperity boasts its status as the "first multidisciplinary aggregator of OA journals and papers." Their focus is on helping you avoid paywalls while connecting you to authoritative research. In addition to providing readers with easy access to thousands of journals, Paperity seeks to help authors reach their audiences and help journals increase their exposure to boost readership.
Paperity has journal articles for every discipline, and the database offers more than a dozen advanced search options, including the length of the paper and the number of authors. There is even an option to include, exclude, or exclusively search gray papers.
Paperity is available for mobile, with both a mobile site and the Paperity Reader, an app that is available for both Android and Apple users. The database is also available on social media. You can interact with Paperity via Twitter and Facebook, and links to their social media are available on their homepage, including their Twitter feed.
Collection : 8,837,396
Advanced Search Options : Title, abstract, journal title, journal ISSN, publisher, year of publication, number of characters, number of authors, DOI, author, affiliation, language, country, region, continent, gray papers
13. dblp Computer Science Bibliography
The dblp Computer Science Bibliography is an online index of major computer science publications. dblp was founded in 1993, though until 2010 it was a university-specific database at the University of Trier in Germany. It is currently maintained by the Schloss Dagstuhl – Leibniz Center for Informatics.
Although it provides access to both OA articles and those behind a paywall, you can limit your search to only OA articles. The site indexes more than three million publications, making it an invaluable resource in the world of computer science. dblp entries are color-coded based on the type of item.
dblp has an extensive FAQ section, so questions that might arise about topics like the database itself, navigating the website, or the data on dblp, in addition to several other topics, are likely to be answered. The website also hosts a blog and has a section devoted to website statistics.
Collection : 5,884,702
14. EconBiz
EconBiz is a great resource for economic and business studies. A service of the Leibniz Information Centre for Economics, it offers access to full texts online, with the option of searching for OA material only. Their literature search is performed across multiple international databases.
EconBiz has an incredibly useful research skills section, with resources such as Guided Walk, a service to help students and researchers navigate searches, evaluate sources, and correctly cite references; the Research Guide EconDesk, a help desk to answer specific questions and provide advice to aid in literature searches; and the Academic Career Kit for what they refer to as Early Career Researchers.
Other helpful resources include personal literature lists, a calendar of events for relevant calls for papers, conferences, and workshops, and an economics terminology thesaurus to help in finding keywords for searches. To stay up-to-date with EconBiz, you can sign up for their newsletter.
Collection : 1,075,219
Advanced Search Options : Title, subject, author, institution, ISBN/ISSN, journal, publisher, language, OA only
15. BioMed Central
BioMed Central provides OA research from more than 300 peer-reviewed journals. While originally focused on resources related to the physical sciences, math, and engineering, BioMed Central has branched out to include journals that cover a broader range of disciplines, with the aim of providing a single platform that provides OA articles for a variety of research needs. You can browse these journals by subject or title, or you can search all articles for your required keyword.
BioMed Central has a commitment to peer-reviewed sources and to the peer review process itself, continually seeking to help and improve the peer review process. They're "committed to maintaining high standards through full and stringent peer review."
Additionally, the website includes resources to assist and support editors as part of their commitment to providing high-quality, peer-reviewed OA articles.
Collection : 507,212
Other Services : BMC administers the International Standard Randomised Controlled Trial Number (ISRCTN) registry. While initially designed for registering clinical trials, since its creation in 2000, the registry has broadened its scope to include other health studies as well.
The registry is recognized by the International Committee of Medical Journal Editors, as well as the World Health Organization (WHO), and it meets the requirements established by the WHO International Clinical Trials Registry Platform.
The study records included in the registry are all searchable and free to access. The ISRCTN registry "supports transparency in clinical research, helps reduce selective reporting of results and ensures an unbiased and complete evidence base."
Advanced Search Options : Author, title, journal, list
A multidisciplinary search engine, JURN provides links to various scholarly websites, articles, and journals that are free to access or OA. Covering the fields of the arts, humanities, business, law, nature, science, and medicine, JURN has indexed almost 5,000 repositories to help you find exactly what you're looking for.
Search features are enhanced by Google, but searches are filtered through their index of repositories. JURN seeks to reach a wide audience, with their search engine tailored to researchers from "university lecturers and students seeking a strong search tool for OA content" and "advanced and ambitious students, age 14-18" to "amateur historians and biographers" and "unemployed and retired lecturers."
That being said, JURN is very upfront about its limitations. They admit to not being a good resource for educational studies, social studies, or psychology, and conference archives are generally not included due to frequently unstable URLs.
Collection : 5,064 indexed journals
Other Services : JURN has a browser add-on called UserScript. This add-on allows users to integrate the JURN database directly into Google Search. When performing a search through Google, the add-on creates a link that sends the search directly to JURN CSE. JURN CSE is a search service that is hosted by Google.
Clicking the link from the Google Search bar will run your search through the JURN database from the Google homepage. There is also an interface for a DuckDuckGo search box; while this search engine has an emphasis on user privacy, for smaller sites that may be indexed by JURN, DuckDuckGo may not provide the same depth of results.
Advanced Search Options : Google search modifiers
Dryad is a digital repository of curated, OA scientific research data. Launched in 2009, it is run by a not-for-profit membership organization, with a community of institutional and publisher members for whom their services have been designed. Members include institutions such as Stanford, UCLA, and Yale, as well as publishers like Oxford University Press and Wiley.
Dryad aims to "promote a world where research data is openly available, integrated with the scholarly literature, and routinely reused to create knowledge." It is free to access for the search and discovery of data. Their user experience is geared toward easy self-depositing, supports Creative Commons licensing, and provides DOIs for all their content.
Note that there is a publishing charge associated if you wish to publish your data in Dryad. When searching datasets, they are accompanied by author information and abstracts for the associated studies, and citation information is provided for easy attribution.
Collection : 44,458
Advanced Search Options : No
Run by the British Library, the E-Theses Online Service (EThOS) allows you to search over 500,000 doctoral theses in a variety of disciplines. All of the doctoral theses available on EThOS have been awarded by higher education institutions in the United Kingdom.
Although some full texts are behind paywalls, you can limit your search to items available for immediate download, either directly through EThOS or through an institution's website. More than half of the records in the database provide access to full-text theses.
EThOS notes that they do not hold all records for all institutions, but they strive to index as many doctoral theses as possible, and the database is constantly expanding, with approximately 3,000 new records added and 2,000 new full-text theses available every month. The availability of full-text theses is dependent on multiple factors, including their availability in the institutional repository and the level of repository development.
Collection : 500,000+
Advanced Search Options : Abstract, author's first name, author's last name, awarding body, current institution, EThOS ID, year, language, qualifications, research supervisor, sponsor/funder, keyword, title
PubMed is a research platform well-known in the fields of science and medicine. It was created and developed by the National Center for Biotechnology Information (NCBI) at the National Library of Medicine (NLM). It has been available since 1996 and offers access to "more than 33 million citations for biomedical literature from MEDLINE, life science journals, and online books."
While PubMed does not provide full-text articles directly, and many full-text articles may be behind paywalls or require subscriptions to access them, when articles are available from free sources, such as through PubMed Central (PMC), those links are provided with the citations and abstracts that PubMed does provide.
PMC, which was established in 2000 by the NLM, is a free full-text archive that includes more than 6,000,000 records. PubMed records link directly to corresponding PMC results. PMC content is provided by publishers and other content owners, digitization projects, and authors directly.
Collection : 33,000,000+
Advanced Search Options : Author's first name, author's last name, identifier, corporation, date completed, date created, date entered, date modified, date published, MeSH, book, conflict of interest statement, EC/RN number, editor, filter, grant number, page number, pharmacological action, volume, publication type, publisher, secondary source ID, text, title, abstract, transliterated title
20. Semantic Scholar
A unique and easy-to-use resource, Semantic Scholar defines itself not just as a research database but also as a "search and discovery tool." Semantic Scholar harnesses the power of artificial intelligence to efficiently sort through millions of science-related papers based on your search terms.
Through this singular application of machine learning, Semantic Scholar expands search results to include topic overviews based on your search terms, with the option to create an alert for or further explore the topic. It also provides links to related topics.
In addition, search results produce "TLDR" summaries in order to provide concise overviews of articles and enhance your research by helping you to navigate quickly and easily through the available literature to find the most relevant information. According to the site, although some articles are behind paywalls, "the data [they] have for those articles is limited," so you can expect to receive mostly full-text results.
Collection : 203,379,033
Other Services : Semantic Scholar supports multiple popular browsers. Content can be accessed through both mobile and desktop versions of Firefox, Microsoft Edge, Google Chrome, Apple Safari, and Opera.
Additionally, Semantic Scholar provides browser extensions for both Chrome and Firefox, so AI-powered scholarly search results are never more than a click away. The mobile interface includes an option for Semantic Swipe, a new way of interacting with your research results.
There are also beta features that can be accessed as part of the Beta Program, which will provide you with features that are being actively developed and require user feedback for further improvement.
Advanced Search Options : Field of study, date range, publication type, author, journal, conference, PDF
Zenodo, powered by the European Organization for Nuclear Research (CERN), was launched in 2013. Taking its name from Zenodotus, the first librarian of the ancient library of Alexandria, Zenodo is a tool "built and developed by researchers, to ensure that everyone can join in open science." Zenodo accepts all research from every discipline in any file format.
However, Zenodo also curates uploads and promotes peer-reviewed material that is available through OA. A DOI is assigned to everything that is uploaded to Zenodo, making research easily findable and citable. You can sort by keyword, title, journal, and more and download OA documents directly from the site.
While there are closed access and restricted access items in the database, the vast majority of research is OA material. Search results can be filtered by access type, making it easy to view the free articles available in the database.
Collection : 2,220,000+
Advanced Search Options : Access, file type, keywords
Check out our roundup of free research databases as a handy one-page PDF.
How to find peer-reviewed articles.
There are a lot of free scholarly articles available from various sources. The internet is a big place. So how do you go about finding peer-reviewed articles when conducting your research? It's important to make sure you are using reputable sources.
The first source of the article is the person or people who wrote it. Checking out the author can give you some initial insight into how much you can trust what you’re reading. Looking into the publication information of your sources can also indicate whether the article is reliable.
Aspects of the article, such as subject and audience, tone, and format, are other things you can look at when evaluating whether the article you're using is valid, reputable, peer-reviewed material. So, let's break that down into various components so you can assess your research to ensure that you're using quality articles and conducting solid research.
Check the Author
Peer-reviewed articles are written by experts or scholars with experience in the field or discipline they're writing about. The research in a peer-reviewed article has to pass a rigorous evaluation process, so it's a foregone conclusion that the author(s) of a peer-reviewed article should have experience or training related to that research.
When evaluating an article, take a look at the author's information. What credentials does the author have to indicate that their research has scholarly weight behind it? Finding out what type of degree the author has—and what that degree is in—can provide insight into what kind of authority the author is on the subject.
Something else that might lend credence to the author's scholarly role is their professional affiliation. A look at what organization or institution they are affiliated with can tell you a lot about their experience or expertise. Where were they trained, and who is verifying their research?
Identify Subject and Audience
The ultimate goal of a study is to answer a question. Scholarly articles are also written for scholarly audiences, especially articles that have gone through the peer review process. This means that the author is trying to reach experts, researchers, academics, and students in the field or topic the research is based on.
Think about the question the author is trying to answer by conducting this research, why, and for whom. What is the subject of the article? What question has it set out to answer? What is the purpose of finding the information? Is the purpose of the article of importance to other scholars? Is it original content?
Research should also be approached analytically. Is the methodology sound? Is the author using an analytical approach to evaluate the data that they have obtained? Are the conclusions they've reached substantiated by their data and analysis? Answering these questions can reveal a lot about the article's validity.
Format Matters
Reliable articles from peer-reviewed sources have certain format elements to be aware of. The first is an abstract. An abstract is a short summary or overview of the article. Does the article have an abstract? It's unlikely that you're reading a peer-reviewed article if it doesn't. Peer-reviewed journals will also have a word count range. If an article seems far too short or incredibly long, that may be reason to doubt it.
Another feature of reliable articles is the sections the information is divided into. Peer-reviewed research articles will have clear, concise sections that appropriately organize the information. This might include a literature review, methodology, results (in the case of research articles), and a conclusion.
One of the most important sections is the references or bibliography. This is where the researcher lists all the sources of their information. A peer-reviewed source will have a comprehensive reference section.
An article that has been written to reach an academic community will have an academic tone. The language that is used, and the way this language is used, is important to consider. If the article is riddled with grammatical errors, confusing syntax, and casual language, it almost definitely didn't make it through the peer review process.
Also consider the use of terminology. Every discipline is going to have standard terminology or jargon that can be used and understood by other academics in the discipline. The language in a peer-reviewed article is going to reflect that.
If the author is going out of their way to explain simple terms, or terms that are standard to the field or discipline, it's unlikely that the article has been peer reviewed, as this is something that the author would be asked to address during the review process.
Publication
The source of the article will be a very good indicator of the likelihood that it was peer reviewed. Where was the article published? Was it published alongside other academic articles in the same discipline? Is it a legitimate and reputable scholarly publication?
A trade publication or newspaper might be legitimate or reputable, but it is not a scholarly source, and it will not have been subject to the peer review process. Scholarly journals are the best resource for peer-reviewed articles, but it's important to remember that not all scholarly journals are peer reviewed.
It's helpful to look at a scholarly source's website, as peer-reviewed journals will have a clear indication of the peer review process. University libraries, institutional repositories, and reliable databases (and now you have a list of legit ones) can also help provide insight into whether an article comes from a peer-reviewed journal.

Common Research Mistakes to Avoid
Research is a lot of work. Even with high standards and good intentions, it's easy to make mistakes. Perhaps you searched for access to scientific journals for free and found the perfect peer-reviewed sources, but you forgot to document everything, and your references are a mess. Or, you only searched for free online articles and missed out on a ground-breaking study that was behind a paywall.
Whether your research is for a degree or to get published or to satisfy your own inquisitive nature, or all of the above, you want all that work to produce quality results. You want your research to be thorough and accurate.
To have any hope of contributing to the literature on your research topic, your results need to be high quality. You might not be able to avoid every potential mistake, but here are some that are both common and easy to avoid.
Sticking to One Source
One of the hallmarks of good research is a healthy reference section. Using a variety of sources gives you a better answer to your question. Even if all of the literature is in agreement, looking at various aspects of the topic may provide you with an entirely different picture than you would have if you looked at your research question from only one angle.
Not Documenting Every Fact
As you conduct your research, do yourself a favor and write everything down. Everything you include in your paper or article that you got from another source is going to need to be added to your references and cited.
It's important, especially if your aim is to conduct ethical, high-quality research, that all of your research has proper attribution. If you don't document as you go, you could end up making a lot of work for yourself if the information you don't write down is something that later, as you write your paper, you really need.
Using Outdated Materials
Academia is an ever-changing landscape. What was true in your academic discipline or area of research ten years ago may have since been disproven. If fifteen studies have come out since the article that you're using was published, it's more than a little likely that you're going to be basing your research on flawed or dated information.
If the information you're basing your research on isn't as up-to-date as possible, your research won't be of quality or able to stand up to any amount of scrutiny. You don't want all of your hard work to be for naught.
Relying Solely on Open Access Journals
OA is a great resource for conducting academic research. There are high-quality journal articles available through OA, and that can be very helpful for your research. But, just because you have access to free articles, that doesn't mean that there's nothing to be found behind a paywall.
Just as dismissing high-quality peer-reviewed articles because they are OA would be limiting, not exploring any paid content at all is equally short-sighted. If you're seeking to conduct thorough and comprehensive research, exploring all of your options for quality sources is going to be to your benefit.
Digging Too Deep or Not Deep Enough
Research is an art form, and it involves a delicate balance of information. If you conduct your research using only broad search terms, you won't be able to answer your research question well, or you'll find that your research provides information that is closely related to your topic but, ultimately, your findings are vague and unsubstantiated.
On the other hand, if you delve deeply into your research topic with specific searches and turn up too many sources, you might have a lot of information that is adjacent to your topic but without focus and perhaps not entirely relevant. It's important to answer your research question concisely but thoroughly.
Different Types of Scholarly Articles
Different types of scholarly articles have different purposes. An original research article, also called an empirical article, is the product of a study or an experiment. This type of article seeks to answer a question or fill a gap in the existing literature.
Research articles will have a methodology, results, and a discussion of the findings of the experiment or research and typically a conclusion.
Review articles overview the current literature and research and provide a summary of what the existing research indicates or has concluded. This type of study will have a section for the literature review, as well as a discussion of the findings of that review. Review articles will have a particularly extensive reference or bibliography section.
Theoretical articles draw on existing literature to create new theories or conclusions, or look at current theories from a different perspective, to contribute to the foundational knowledge of the field of study.
10 Tips for Navigating Journal Databases
Use the right academic journal database for your search, be that interdisciplinary or specific to your field. Or both!
If it's an option, set the search results to return only peer-reviewed sources.
Start by using search terms that are relevant to your topic without being overly specific.
Try synonyms, especially if your keywords aren't returning the desired results.

Even if you've found some good articles, try searching using different terms.
Explore the advanced search features of the database(s).
Learn to use Booleans (AND, OR, NOT) to expand or narrow your results.
Once you've gotten some good results from a more general search, try narrowing your search.
Read through abstracts when trying to find articles relevant to your research.
Keep track of your research and use citation tools. It'll make life easier when it comes time to compile your references.
7 Frequently Asked Questions
1. how do i get articles for free.
Free articles can be found through free online academic journals, OA databases, or other databases that include OA journals and articles. These resources allow you to access free papers online so you can conduct your research without getting stuck behind a paywall.
Academics don't receive payment for the articles they contribute to journals. There are often, in fact, publication fees that scholars pay in order to publish. This is one of the funding structures that allows OA journals to provide free content so that you don't have to pay fees or subscription costs to access journal articles.
2. How Do I Find Journal Articles?
Journal articles can be found in databases and institutional repositories that can be accessed at university libraries. However, online research databases that contain OA articles are the best resource for getting free access to journal articles that are available online.
Peer-reviewed journal articles are the best to use for academic research, and there are a number of databases where you can find peer-reviewed OA journal articles. Once you've found a useful article, you can look through the references for the articles the author used to conduct their research, and you can then search online databases for those articles, too.
3. How Do I Find Peer-Reviewed Articles?
Peer-reviewed articles can be found in reputable scholarly peer-reviewed journals. High-quality journals and journal articles can be found online using academic search engines and free research databases. These resources are excellent for finding OA articles, including peer-reviewed articles.
OA articles are articles that can be accessed for free. While some scholarly search engines and databases include articles that aren't peer reviewed, there are also some that provide only peer-reviewed articles, and databases that include non-peer-reviewed articles often have advanced search features that enable you to select "peer review only." The database will return results that are exclusively peer-reviewed content.
4. What Are Research Databases?
A research database is a list of journals, articles, datasets, and/or abstracts that allows you to easily search for scholarly and academic resources and conduct research online. There are databases that are interdisciplinary and cover a variety of topics.
For example, Paperity might be a great resource for a chemist as well as a linguist, and there are databases that are more specific to a certain field. So, while ERIC might be one of the best educational databases available for OA content, it's not going to be one of the best databases for finding research in the field of microbiology.
5. How Do I Find Scholarly Articles for Specific Fields?
There are interdisciplinary research databases that provide articles in a variety of fields, as well as research databases that provide articles that cater to specific disciplines. Additionally, a journal repository or index can be a helpful resource for finding articles in a specific field.
When searching an interdisciplinary database, there are frequently advanced search features that allow you to narrow the search results down so that they are specific to your field. Selecting "psychology" in the advanced search features will return psychology journal articles in your search results. You can also try databases that are specific to your field.
If you're searching for law journal articles, many law reviews are OA. If you don't know of any databases specific to history, visiting a journal repository or index and searching "history academic journals" can return a list of journals specific to history and provide you with a place to begin your research.
6. Are Peer-Reviewed Articles Really More Legitimate?
The short answer is yes, peer-reviewed articles are more legitimate resources for academic research. The peer review process provides legitimacy, as it is a rigorous review of the content of an article that is performed by scholars and academics who are experts in their field of study. The review provides an evaluation of the quality and credibility of the article.
Non-peer-reviewed articles are not subject to a review process and do not undergo the same level of scrutiny. This means that non-peer-reviewed articles are unlikely, or at least not as likely, to meet the same standards that peer-reviewed articles do.
7. Are Free Article Directories Legitimate?
Yes! As with anything, some databases are going to be better for certain requirements than others. But, a scholarly article database being free is not a reason in itself to question its legitimacy.
Free scholarly article databases can provide access to abstracts, scholarly article websites, journal repositories, and high-quality peer-reviewed journal articles. The internet has a lot of information, and it's often challenging to figure out what information is reliable.
Research databases and article directories are great resources to help you conduct your research. Our list of the best research paper websites is sure to provide you with sources that are totally legit.
Get Professional Academic Editing
Hire an expert academic editor , or get a free sample, about the author.

Scribendi's in-house editors work with writers from all over the globe to perfect their writing. They know that no piece of writing is complete without a professional edit, and they love to see a good piece of writing transformed into a great one. Scribendi's in-house editors are unrivaled in both experience and education, having collectively edited millions of words and obtained numerous degrees. They love consuming caffeinated beverages, reading books of various genres, and relaxing in quiet, dimly lit spaces.
Have You Read?
"The Complete Beginner's Guide to Academic Writing"
Related Posts

How to Write a Research Proposal

How to Write a Scientific Paper

How to Write a Thesis or Dissertation
Upload your file(s) so we can calculate your word count, or enter your word count manually.
We will also recommend a service based on the file(s) you upload.
| File | Word Count | Include in Price? |
|---|
English is not my first language. I need English editing and proofreading so that I sound like a native speaker.
I need to have my journal article, dissertation, or term paper edited and proofread, or I need help with an admissions essay or proposal.
I have a novel, manuscript, play, or ebook. I need editing, copy editing, proofreading, a critique of my work, or a query package.
I need editing and proofreading for my white papers, reports, manuals, press releases, marketing materials, and other business documents.
I need to have my essay, project, assignment, or term paper edited and proofread.
I want to sound professional and to get hired. I have a resume, letter, email, or personal document that I need to have edited and proofread.
Prices include your personal % discount.
Prices include % sales tax ( ).

Academia.edu no longer supports Internet Explorer.
To browse Academia.edu and the wider internet faster and more securely, please take a few seconds to upgrade your browser .
Download 55 million PDFs for free
Explore our top research interests.

Engineering

Anthropology

- Earth Sciences

- Computer Science

- Mathematics

- Health Sciences

Join 264 million academics and researchers
Track your impact.
Share your work with other academics, grow your audience and track your impact on your field with our robust analytics
Discover new research
Get access to millions of research papers and stay informed with the important topics around the world
Publish your work
Publish your research with fast and rigorous service through Academia.edu Publishing. Get instant worldwide dissemination of your work
Unlock the most powerful tools with Academia Premium

Work faster and smarter with advanced research discovery tools
Search the full text and citations of our millions of papers. Download groups of related papers to jumpstart your research. Save time with detailed summaries and search alerts.
- Advanced Search
- PDF Packages of 37 papers
- Summaries and Search Alerts

Share your work, track your impact, and grow your audience
Get notified when other academics mention you or cite your papers. Track your impact with in-depth analytics and network with members of your field.
- Mentions and Citations Tracking
- Advanced Analytics
- Publishing Tools
Real stories from real people

Used by academics at over 15,000 universities

Get started and find the best quality research
- Academia.edu Publishing
- We're Hiring!
- Help Center
- Find new research papers in:
- Cognitive Science
- Academia ©2024
🇺🇦 make metadata, not war
A comprehensive bibliographic database of the world’s scholarly literature
The world’s largest collection of open access research papers, machine access to our vast unique full text corpus, core features, indexing the world’s repositories.
We serve the global network of repositories and journals
Comprehensive data coverage
We provide both metadata and full text access to our comprehensive collection through our APIs and Datasets
Powerful services
We create powerful services for researchers, universities, and industry
Cutting-edge solutions
We research and develop innovative data-driven and AI solutions
Committed to the POSI
Cost-free PIDs for your repository
OAI identifiers are unique identifiers minted cost-free by repositories. Ensure that your repository is correctly configured, enabling the CORE OAI Resolver to redirect your identifiers to your repository landing pages.
OAI IDs provide a cost-free option for assigning Persistent Identifiers (PIDs) to your repository records. Learn more.
Who we serve?
Enabling others to create new tools and innovate using a global comprehensive collection of research papers.

“ Our partnership with CORE will provide Turnitin with vast amounts of metadata and full texts that we can ... ” Show more
Gareth Malcolm, Content Partner Manager at Turnitin
Academic institutions.
Making research more discoverable, improving metadata quality, helping to meet and monitor open access compliance.

“ CORE’s role in providing a unified search of repository content is a great tool for the researcher and ex... ” Show more
Nicola Dowson, Library Services Manager at Open University
Researchers & general public.
Tools to find, discover and explore the wealth of open access research. Free for everyone, forever.

“ With millions of research papers available across thousands of different systems, CORE provides an invalu... ” Show more
Jon Tennant, Rogue Paleontologist and Founder of the Open Science MOOC
Helping funders to analyse, audit and monitor open research and accelerate towards open science.

“ Aggregation plays an increasingly essential role in maximising the long-term benefits of open access, hel... ” Show more
Ben Johnson, Research Policy Adviser at Research England
Our services, access to raw data.
Create new and innovative solutions.
Content discovery
Find relevant research and make your research more visible.
Managing content
Manage how your research content is exposed to the world.
Companies using CORE

Gareth Malcolm
Content Partner Manager at Turnitin
Our partnership with CORE will provide Turnitin with vast amounts of metadata and full texts that we can utilise in our plagiarism detection software.
Academic institution using CORE
Kathleen Shearer
Executive Director of the Confederation of Open Access Repositories (COAR)
CORE has significantly assisted the academic institutions participating in our global network with their key mission, which is their scientific content exposure. In addition, CORE has helped our content administrators to showcase the real benefits of repositories via its added value services.
Partner projects

Ben Johnson
Research Policy Adviser
Aggregation plays an increasingly essential role in maximising the long-term benefits of open access, helping to turn the promise of a 'research commons' into a reality. The aggregation services that CORE provides therefore make a very valuable contribution to the evolving open access environment in the UK.

15 Best Websites to Download Research Papers for Free

Is your thirst for knowledge limited by expensive subscriptions? Explore the best websites to download research papers for free and expand your academic reach.
With paywalls acting like impenetrable fortresses, accessing scholarly articles becomes a herculean task. However, a beacon of hope exists in the form of free-access platforms, quenching our thirst for intellectual wisdom. Let’s set sail on this scholarly journey.
Table of Contents
Today’s champions of academia aren’t just about offering free access, they uphold ethics and copyright respectability. Let’s delve into these repositories that are reshaping the academia world. You can download free research papers from any of the following websites.

Best Websites to Download Research Papers
#1. sci-hub – best for accessing paywalled academic papers.
Despite its contentious standing, Sci-Hub offers an invaluable service to knowledge-seekers. While navigating the tightrope between access and legality, it represents a game-changing force in the world of academic research.
#2. Library Genesis (Libgen) – Best for a Wide Range of Books and Articles
It’s not just a repository, but a vibrant confluence of multiple disciplines and interests, catering to the unique intellectual appetite of each knowledge seeker.
What are the benefits of Libgen?
Source: https://libgen.is
#3. Unpaywall – Best for Legal Open Access Versions of Scholarly Articles
What are the benefits of Unpaywall?
#4. Directory of Open Access Journals (DOAJ) – Best for Peer-Reviewed Open Access Journals
Source: https://doaj.org
#5. Open Access Button – Best for Free Versions of Paywalled Articles
What are the benefits of Open Access Button?
#6. Science Open – Best for a Wide Variety of Open Access Scientific Research
Consider Science Open as a bustling town square in the city of scientific knowledge, where scholars from all walks of life gather, discuss, and dissect over 60 million articles.
You might also like:
#7. CORE – Best for Open Access Content Across Disciplines
With its unparalleled aggregation and comprehensive access, CORE embodies the grand orchestra of global research. It stands as an essential tool in the modern researcher’s toolkit.
#8. ERIC – Best for Education Research
What are the benefits of ERIC?
#9. PaperPanda – Best for Free Access to Research Papers
It’s like having a personal research assistant, guiding you through the maze of scholarly literature.
#10. Citationsy Archives – Best for Research Papers from Numerous Fields
Source: https://citationsy.com
#11. OA.mg – Best for Direct Download Links to Open Access Papers
Source: https://oa.mg
#12. Social Science Research Network (SSRN) – Best for Social Sciences and Humanities Research
SSRN serves as an invaluable resource for researchers in the social sciences and humanities, fostering a community that drives innovation and advancements in these fields.
#13. Project Gutenberg – Best for Free Access to eBooks
Project Gutenberg serves as a testament to the power of literature and the accessibility of knowledge. It enables readers worldwide to embark on intellectual journeys through its extensive collection of free eBooks.
#14. PLOS (Public Library of Science) – Best for Open Access to Scientific and Medical Research
As a leading publisher of open access research, PLOS fosters the dissemination of cutting-edge scientific discoveries to a global audience.
#15. arXiv.org – Best for Preprints in Science, Mathematics, and Computer Science
In a world where knowledge is king, accessing a research paper shouldn’t feel like an impossible task. Thanks to these free and innovative websites, we can escape the barriers of paywalls and dive into a vast ocean of intellectual wealth.
Leave a Comment Cancel reply
This website uses cookies to ensure you get the best experience. Learn more about DOAJ’s privacy policy.
Hide this message
You are using an outdated browser. Please upgrade your browser to improve your experience and security.
The Directory of Open Access Journals
Directory of Open Access Journals
Find open access journals & articles.
Doaj in numbers.
80 languages
134 countries represented
13,573 journals without fees
20,555 journals
10,200,699 article records
Quick search
About the directory.
DOAJ is a unique and extensive index of diverse open access journals from around the world, driven by a growing community, and is committed to ensuring quality content is freely available online for everyone.
DOAJ is committed to keeping its services free of charge, including being indexed, and its data freely available.
→ About DOAJ
→ How to apply
DOAJ is twenty years old in 2023.
Fund our 20th anniversary campaign
DOAJ is independent. All support is via donations.
82% from academic organisations
18% from contributors
Support DOAJ
Publishers don't need to donate to be part of DOAJ.
News Service
Vacancy: operations manager, press release: pubscholar joins the movement to support the directory of open access journals, new major version of the api to be released, toutes les pages de notre «guide to applying» sont désormais disponibles en français, all pages in our guide to applying are now available in french, the story behind the journal: sortuz.
→ All blog posts
We would not be able to work without our volunteers, such as these top-performing editors and associate editors.
→ Meet our volunteers
Librarianship, Scholarly Publishing, Data Management
Brisbane, Australia (Chinese, English)
Adana, Türkiye (Turkish, English)
Humanities, Social Sciences
Natalia Pamuła
Toruń, Poland (Polish, English)
Medical Sciences, Nutrition
Pablo Hernandez
Caracas, Venezuela (Spanish, English)
Research Evaluation
Paola Galimberti
Milan, Italy (Italian, German, English)
Social Sciences, Humanities
Dawam M. Rohmatulloh
Ponorogo, Indonesia (Bahasa Indonesia, English, Dutch)
Systematic Entomology
Kadri Kıran
Edirne, Türkiye (English, Turkish, German)
Library and Information Science
Nataliia Kaliuzhna
Kyiv, Ukraine (Ukrainian, Russian, English, Polish)
Recently-added journals
DOAJ’s team of managing editors, editors, and volunteers work with publishers to index new journals. As soon as they’re accepted, these journals are displayed on our website freely accessible to everyone.
→ See Atom feed
→ A log of journals added (and withdrawn)
→ DOWNLOAD all journals as CSV
- Maǧallaẗ Markaz Al-Buḥūṯ Al-Ǧuġrāfiyyaẗ wa Al-Kārtūğirāfiyyaẗ
- المجلة المصرية للدراسات المتخصصة
- Maǧallaẗ Buḥūṯ Al-Šarq Al-Awsaṭ
- Advanced Spine Journal
- فصلنامه ژئوپلتیک
- Jornal de Assistência Farmacêutica e Farmacoeconomia
- Informatische Bildung in Schulen
- Studia Filmoznawcze
- Journal of Rare Diseases
- پدافند غیرعامل
- Electromagnetic Science
- Online Media and Global Communication
- Česká Stomatologie a Praktické Zubní Lékařství
- Configurazioni
- Frontiers in Arachnid Science
WeChat QR code

- Interesting
- Scholarships
- UGC-CARE Journals
14 Websites to Download Research Paper for Free – 2024
14 Top Websites for Free, Open Access Research Paper Download
Collecting and reading relevant research articles to one’s research areas is important for PhD scholars. However, for any research scholar, downloading a research paper is one of the most difficult tasks. You must pay for access to high-quality research materials or subscribe to the journal or publication. In this article, ilovephd lists the top 14 websites to download free research papers, journals, books, datasets, patents, and conference proceedings downloads.
Free Research Paper Download Websites – 2024
Check the 14 best free websites to download and read research papers listed below:
Sci-Hub is a website link with over 64.5 million academic papers and articles available for direct download. It bypasses publisher paywalls by allowing access through educational institution proxies. To download papers Sci-Hub stores papers in its repository, this storage is called Library Genesis (LibGen) or Library Genesis Proxy 2024. It helps researchers to download free articles by simply using Digital Object Identifier (DOI) of the article.

Visit: Working Sci-Hub Proxy Links – 2024
2. Z-Library
The Z-Library clones Library Genesis, a shadow library project. Z-Library facilitates file sharing of scholarly journal articles, academic texts, and general-interest books (including some copyrighted materials). While most of its books come from Library Genesis, further expanding the collection, users can also directly upload content to the site. This user-contributed content helps to make literature even more widely available. Additionally, individuals can donate to the website’s repository, furthering their mission of free access.
Z-Library claims to have a massive collection, boasting more than 10,139,382 Books books and 84,837,646 Articles articles as of April 25, 2024. According to the project’s page for academic publications (at booksc.org), it aspires to be “the world’s largest e-book library” as well as “the world’s largest scientific papers repository.” Interestingly, Z-Library also describes itself as a donation-based non-profit organization.

Visit Z-Library – You can Download 70,000,000+ scientific articles for free
3. Library Genesis
The Library Genesis aggregator is a community aiming to collect and catalog item descriptions for the most scientific, scientific, and technical directions, as well as file metadata. In addition to the descriptions, the aggregator contains only links to third-party resources hosted by users. All information posted on the website is collected from publicly available public Internet resources and is intended solely for informational purposes.
Visit: libgen.li
4. Unpaywall – Free Research Paper Download
Unpaywall harvests Open Access content from over 50,000 publishers and repositories, and makes it easy to find, track, and use. It is integrated into thousands of library systems, search platforms, and other information products worldwide. If you’re involved in scholarly communication, there’s a good chance you’ve already used Unpaywall data.
Unpaywall is run by OurResearch, a nonprofit dedicated to making scholarships more accessible to everyone. Open is our passion. So it’s only natural our source code is open, too.

Visit: unpaywall.org
5. GetTheResearch.org
GetTheResearch.org is an Artificial Intelligence(AI) powered search engine for searching and understanding scientific articles for researchers and scientists. It was developed as a part of the Unpaywall project. Unpaywall is a database of 23,329,737 free scholarly Open Access(OA) articles from over 50,000 publishers and repositories, and make it easy to find, track, and use.

Visit: Find and Understand 25 Million Peer-Reviewed Research Papers for Free
6. Directory of Open Access Journals (DOAJ)
DOAJ (Directory of Open Access Journals) was launched in 2003 with 300 open-access journals. Today, this independent index contains almost 17,500 peer-reviewed, open-access journals covering all areas of science, technology, medicine, social sciences, arts, and humanities. Open-access journals from all countries and in all languages are accepted for indexing.
DOAJ is financially supported by many libraries, publishers, and other like-minded organizations. Supporting DOAJ demonstrates a firm commitment to open access and the infrastructure that supports it.
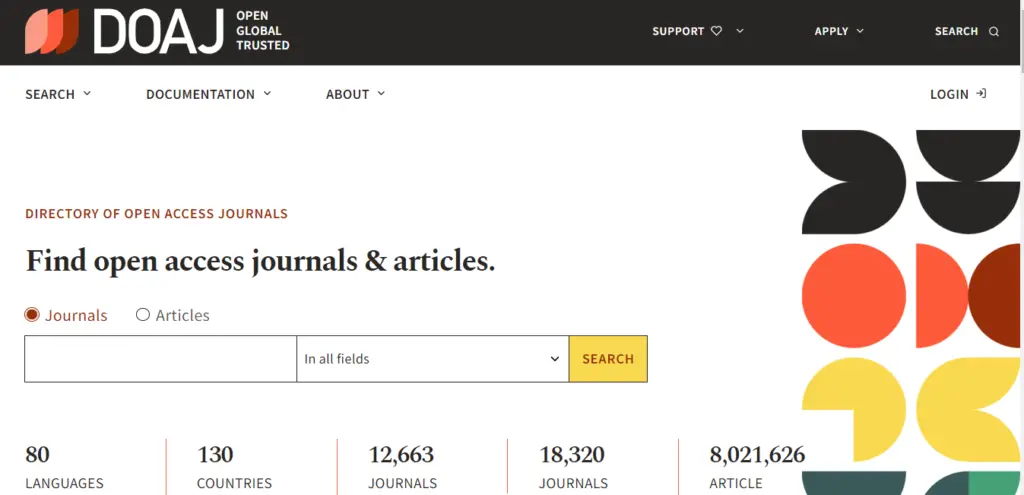
Visit: doaj.org
7. Researcher
The Researcher is a free journal-finding mobile application that helps you to read new journal papers every day that are relevant to your research. It is the most popular mobile application used by more than 3 million scientists and researchers to keep themselves updated with the latest academic literature.
Visit: 10 Best Apps for Graduate Students
8. Science Open
ScienceOpen is a discovery platform with interactive features for scholars to enhance their research in the open, make an impact, and receive credit for it. It provides context-building services for publishers, to bring researchers closer to the content than ever before. These advanced search and discovery functions, combined with post-publication peer review, recommendation, social sharing, and collection-building features make ScienceOpen the only research platform you’ll ever need.
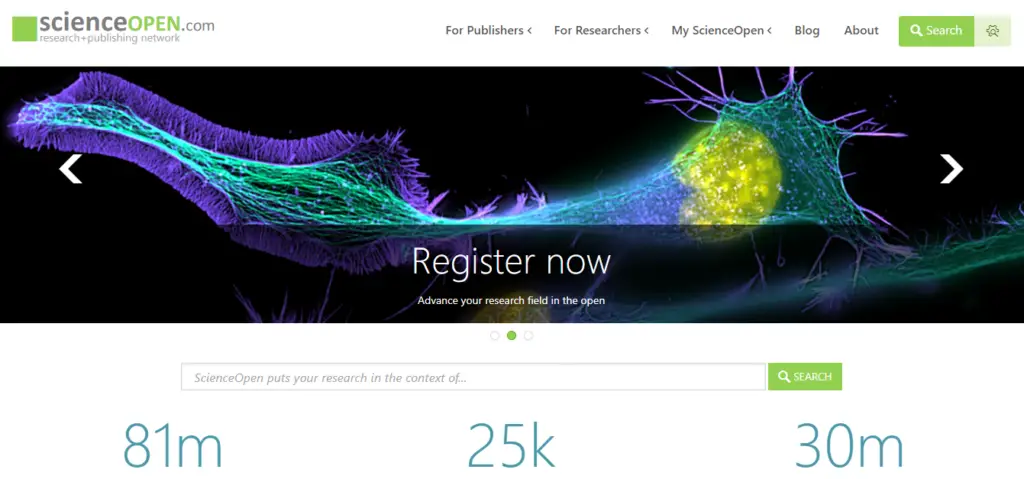
Visit: scienceopen.com
OA.mg is a search engine for academic papers. Whether you are looking for a specific paper, or for research from a field, or all of an author’s works – OA.mg is the place to find it.
Visit: oa.mg
10. Internet Archive Scholar
Internet Archive Scholar (IAS) is a full-text search index that includes over 25 million research articles and other scholarly documents preserved in the Internet Archive. The collection spans from digitized copies of eighteenth-century journals through the latest Open Access conference proceedings and pre-prints crawled from the World Wide Web.

Visit: Sci hub Alternative – Internet Archive Scholar
11. Citationsy Archives
Citationsy was founded in 2017 after the reference manager Cenk was using at the time, RefMe, was shut down. It was immediately obvious that the reason people loved RefMe — a clean interface, speed, no ads, and simplicity of use — did not apply to CiteThisForMe. It turned out to be easier than anticipated to get a rough prototype up.

Visit: citationsy.com
CORE is the world’s largest aggregator of open-access research papers from repositories and journals. It is a not-for-profit service dedicated to the open-access mission. We serve the global network of repositories and journals by increasing the discoverability and reuse of open-access content.
It provides solutions for content management, discovery, and scalable machine access to research. Our services support a wide range of stakeholders, specifically researchers, the general public, academic institutions, developers, funders, and companies from a diverse range of sectors including but not limited to innovators, AI technology companies, digital library solutions, and pharma.

Visit: core.ac.uk
13. Dimensions
The database called “Dimensions” covers millions of research publications connected by more than 1.6 billion citations, supporting grants, datasets, clinical trials, patents, and policy documents.
Dimensions is the most comprehensive research grants database that links grants to millions of resulting publications, clinical trials, and patents. It
provides up-to-the-minute online attention data via Altmetric, showing you how often publications and clinical trials are discussed around the world. 226m Altmetric mentions with 17m links to publications.
Dimensions include datasets from repositories such as Figshare, Dryad, Zenodo, Pangaea, and many more. It hosts millions of patents with links to other citing patents as well as to publications and supporting grants.
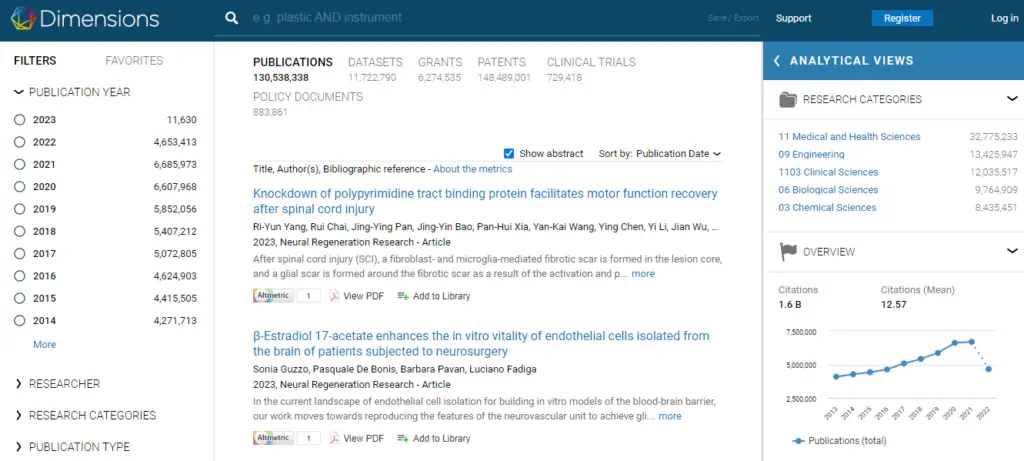
Visit: dimensions.ai
14. PaperPanda – Download Research Papers for Free
PaperPanda is a Chrome extension that uses some clever logic and the Panda’s detective skills to find you the research paper PDFs you need. Essentially, when you activate PaperPanda it finds the DOI of the paper from the current page, and then goes and searches for it. It starts by querying various Open Access repositories like OpenAccessButton, OaDoi, SemanticScholar, Core, ArXiV, and the Internet Archive. You can also set your university library’s domain in the settings (this feature is in the works and coming soon). PaperPanda will then automatically search for the paper through your library. You can also set a different custom domain in the settings.
Visit: PaperPanda
I hope this article will help you to know some of the best websites to download research papers and journals for free. By utilizing open-access databases, free search tools, and potentially even your local university library, you can access a wealth of valuable scholarly information without infringing on a copyright. Remember, ethical practices in research paper downloading are important, so always prioritize legal access to materials whenever possible. Happy researching!

- download paid books for free
- download research papers for free
- download research papers free
- download scientific article for free
- Free Datasets download
- how to download research paper
Top 7 Artificial Intelligence (AI) Tools in Scientific Research 2024
Reviewer three: unveiling the world of peer review, quantitative vs qualitative research.
hi im zara,student of art. could you please tell me how i can download the paper and books about painting, sewing,sustainable fashion,graphic and so on. thank a lot
thanks for the informative reports.
warm regards
Good, Keep it up!
LEAVE A REPLY Cancel reply
Most popular, india – sri lanka joint research funding opportunity, india-eu partner up for explainable and robust ai research, swiss government excellence scholarships, top 100 machine learning topics and 10 research ideas – 2025, auto-gpt: the next-level ai tool surpassing chatgpt for complex tasks, 40 part-time jobs websites for phd scholars to earn extra income, advantages and disadvantages of getting a patent, best for you, 24 best online plagiarism checker free – 2024, what is phd, popular posts, how to check scopus indexed journals 2024, scopus indexed journals list 2024, popular category.
- POSTDOC 317
- Interesting 258
- Journals 234
- Fellowship 131
- Research Methodology 102
- All Scopus Indexed Journals 92
Mail Subscription

iLovePhD is a research education website to know updated research-related information. It helps researchers to find top journals for publishing research articles and get an easy manual for research tools. The main aim of this website is to help Ph.D. scholars who are working in various domains to get more valuable ideas to carry out their research. Learn the current groundbreaking research activities around the world, love the process of getting a Ph.D.
Contact us: [email protected]
Google News
Copyright © 2024 iLovePhD. All rights reserved
- Artificial intelligence

- svg]:stroke-accent-900">
How to get past paywalls and read scientific studies
By Whitson Gordon
Posted on Oct 23, 2019 9:37 PM EDT
4 minute read
Popular Science stories often link directly to scientific studies. You can get all the information you need from the articles themselves, and even more from these links, but if you get the urge to investigate further—perhaps to see the data for yourself—you’ll want to read the study firsthand. Unfortunately, many academic papers are hidden behind expensive paywalls.
There’s a lot to say about the academic research industry, but many believe scientific studies should be freely available to the public . Even if you find a paper that’s hidden behind a paid subscription, there are ways to get it for free—and we’re not talking about piracy. Often, the study you’re looking for may be freely, legally available elsewhere, if you know how to find it.
Google (Scholar) it
Don’t get discouraged just because one database says you need to pay for a specific study. Search for the title of the study (or a portion of the title with an author’s last name) on Google Scholar , the Google-powered search engine for academic literature. If you’re lucky, you’ll see a result with an [html] or [pdf] link on the right-hand side of the page, which should link you to the full text of the study.
If for some reason the right sidebar link doesn’t work, you can also click the “All 11 versions” link at the bottom of each result block to see more sites that offer the paper. You could also try searching regular ol’ Google for the paper’s title, perhaps with the filetype:PDF operator as part of your search terms. This may help you find it on sites that aren’t crawled by Google Scholar.
Use browser extensions to your advantage
If you’re a journalist, student, or science nerd who finds yourself regularly hunting for full-text articles, you can streamline the process a bit with a browser extension called Unpaywall . It works with Google Chrome and Mozilla Firefox, and displays a small padlock icon on the right side of your browser window whenever you visit a page dedicated to a scholarly article. If there’s a paywall and the padlock is green, that means Unpaywall found a free version somewhere on the web, and you can click the icon to visit it immediately. In my experience, this doesn’t find much more beyond a Google Scholar search, but it’s a lot easier than performing manual searches all the time. Heck, even if a site isn’t paywalled, Unpaywall’s green icon is still easier than hunting for the “Download PDF” button on a given page.
A tool called Open Access Button does something similar. It’s browser-agnostic and has been around for a bit longer, so try both tools and see which you like better. I think Unpaywall feels a bit smoother, but Open Access Button’s maturity may help you find things Unpaywall doesn’t know about yet.
Check your local library
Many public libraries subscribe to academic databases and share those subscriptions with their constituents. You may have to head to the library’s physical location to get a library card, if you don’t have one already, but those are usually free or cheap. And from then on, you should be able to access a lot of your library’s resources right from your computer at home, including scholarly journals (not to mention other paywalled magazines like Consumer Reports). If your library doesn’t have access to the publication you’re looking for, they may even be able to get it through an inter-library loan . If you ever feel lost, don’t hesitate to ask a librarian—they probably know the process like the back of their hand, and will do their best to help you find what you’re looking for.
If you’re a student, your school or university likely has access to more databases than you can shake a stick at (not to mention hordes of physical journals you can hunt through). If you aren’t a student but have a college nearby, ask them if they offer fee-based library cards—you may be able to pay for in-house access to their vast resources.
Email the study’s author
Finally, if you can’t find a paper anywhere online, you might be able to get it directly from one of the people who wrote it. The money earned by those paywalls doesn’t go to the researchers—it goes to the publisher, so authors are often happy to give you a copy of their paper for free (provided they’re allowed to do so).
Finding their current email address is the hard part. Papers will often contain an email address you can contact for questions, but if this becomes out of date, you’ll have to do a little hunting. Find the university or organization the researcher currently works for, not the one they worked for when the study was first published. A little Googling can usually point you in the right direction, but sites like ResearchGate and LinkedIn can also help. Some researchers have a personal website that may be up to date, as well, and in some cases, may even have their previous work available to download. But if not, shoot them a message, ask politely if they’d be willing to send you a copy, and thank them for their hard work.
Latest in Tech Hacks
How to customize screensavers on your tv how to customize screensavers on your tv.
By David Nield
How to use LinkedIn AI tools to find a job How to use LinkedIn AI tools to find a job

- About the Centre
- Conferences
- School of Economics
NICEP 2024-12: Foreign influence in US politics
This paper documents that foreign lobbying influences US government spending. We introduce a comprehensive dataset of over 230,000 date-stamped, in-person meetings between agents representing foreign governments and individual US legislators, state governors, and employees of US executive agencies from 2000 to 2018. The data suggest that foreign agents meet disproportionately with individuals important for foreign aid and corporate subsidies, like legislators sitting on powerful congressional committees. Foreign agents also maintain connections with legislators even after they depart powerful committees, providing evidence that meetings do not just reflect short-term quid-pro-quo arrangements. Around meetings, foreign countries receive greater amounts of financial aid. Foreign firms whose governments lobby more often also receive larger corporate subsidies from areas the legislators and governors that they meet with represent. Finally, legislators who meet more often with foreign agents receive both monetary and electoral benefits, while we do not find changes in the political contributions they receive or in their probability of re-election, suggesting that legislators are not punished by their constituents for meeting with representatives of foreign countries.
Download the paper in PDF format
Marco Grotteria, Max Miller and S.Lakshmi Naaraayanan
View all NICEP working papers
Nottingham Interdisciplinary Centre for Economic and Political Research
University of Nottingham Law and Social Sciences Building University Park Nottingham, NG7 2RD
telephone: +44 (0)115 84 68135 email: [email protected]
Legal information
- Terms and conditions
- Posting rules
- Accessibility
- Freedom of information
- Charity gateway
- Cookie policy
Connect with the University of Nottingham through social media and our blogs .
This paper is in the following e-collection/theme issue:
Published on 24.6.2024 in Vol 26 (2024)
Finding Medical Photographs of Patients Online: Randomized, Cross-Sectional Study
Authors of this article:

Original Paper
- Zack Marshall 1, 2 , MSW, PhD ;
- Maushumi Bhattacharjee 3 , LLB, LLM ;
- Meng Wang 1 , PhD ;
- Abdul Cadri 4 , MPH ;
- Hannah James 5 ;
- Shabnam Asghari 6 , MD, MPH, PhD ;
- Rene Peltekian 7 , MSW ;
- Veronica Benz 1 , MSW ;
- Vanessa Finley-Roy 8 , SW, MSc ;
- Brynna Childs 2, 9 , BA, MISt ;
- Lauren Asaad 1 , BHSc ;
- Michelle Swab 10 , MA, MLIS ;
- Vivian Welch 11 , PhD ;
- Fern Brunger 12 , PhD ;
- Chris Kaposy 12 , PhD
1 Department of Community Health Sciences, Cumming School of Medicine, University of Calgary, Calgary, AB, Canada
2 School of Social Work, McGill University, Montreal, QC, Canada
3 Faculty of Law, McGill University, Montreal, QC, Canada
4 Department of Family Medicine, Faculty of Medicine and Health Sciences, McGill University, Montreal, QC, Canada
5 Department of Anatomy & Cell Biology, McGill University, Montreal, QC, Canada
6 Faculty of Medicine, Memorial University of Newfoundland, St John's, NL, Canada
7 Renison University College, University of Waterloo, Waterloo, ON, Canada
8 Faculty of Medicine, Universite de Montreal, Montreal, QC, Canada
9 CKUT 90.3FM Radio McGill, Montreal, QC, Canada
10 Health Sciences Library, Memorial University, St John's, NL, Canada
11 Bruyere Research Institute, Ottawa, ON, Canada
12 Division of Population Health and Applied Health Sciences, Faculty of Medicine, Memorial University, St John's, NL, Canada
Corresponding Author:
Zack Marshall, MSW, PhD
Department of Community Health Sciences
Cumming School of Medicine
University of Calgary
3280 Hospital Drive NW
Calgary, AB, T2N 4Z6
Phone: 1 4032206940
Email: [email protected]
Background: Photographs from medical case reports published in academic journals have previously been found in online image search results. This means that patient photographs circulate beyond the original journal website and can be freely accessed online. While this raises ethical and legal concerns, no systematic study has documented how often this occurs.
Objective: The aim of this cross-sectional study was to provide systematic evidence that patient photographs from case reports published in medical journals appear in Google Images search results. Research questions included the following: (1) what percentage of patient medical photographs published in case reports were found in Google Images search results? (2) what was the relationship between open access publication status and image availability? and (3) did the odds of finding patient photographs on third-party websites differ between searches conducted in 2020 and 2022?
Methods: The main outcome measure assessed whether at least 1 photograph from each case report was found on Google Images when using a structured search. Secondary outcome variables included the image source and the availability of images on third-party websites over time. The characteristics of medical images were described using summary statistics. The association between the source of full-text availability and image availability on Google Images was tested using logistic regressions. Finally, we examined the trend of finding patient photographs using generalized estimating equations.
Results: From a random sample of 585 case reports indexed in PubMed, 186 contained patient photographs, for a total of 598 distinct images. For 142 (76.3%) out of 186 case reports, at least 1 photograph was found in Google Images search results. A total of 18.3% (110/598) of photographs included eye, face, or full body, including 10.9% (65/598) that could potentially identify the patient. The odds of finding an image from the case report online were higher if the full-text paper was available on ResearchGate (odds ratio [OR] 9.16, 95% CI 2.71-31.02), PubMed Central (OR 7.90, 95% CI 2.33-26.77), or Google Scholar (OR 6.07, 95% CI 2.77-13.29) than if the full-text was available solely through an open access journal (OR 5.33, 95% CI 2.31-12.28). However, all factors contributed to an increased risk of locating patient images online. Compared with the search in 2020, patient photographs were less likely to be found on third-party websites based on the 2022 search results (OR 0.61, 95% Cl 0.43-0.87).
Conclusions: A high proportion of medical photographs from case reports was found on Google Images, raising ethical concerns with policy and practice implications. Journal publishers and corporations such as Google are best positioned to develop an effective remedy. Until then, it is crucial that patients are adequately informed about the potential risks and benefits of providing consent for clinicians to publish their images in medical journals.
Introduction
Case reports are an important tool for medical, scientific, and educational purposes [ 1 ]. Written by practicing clinicians, peer-reviewed case reports provide relevant and timely medical information that contributes to evidence-based practice [ 1 , 2 ]. A large number of case reports are published each year; for example, 74,270 case reports were published in 2022 and indexed in PubMed.
Case reports often include images, including patient photographs [ 3 , 4 ]. Guidelines related to the publication of medical photographs in case reports often refer to overarching statements such as the Declaration of Helsinki, or slightly more specific policies such as the guidelines outlined by the Committee on Publication Ethics (COPE), or the Case Report (CARE) guidelines for case reports [ 5 - 7 ]. The Declaration of Helsinki states that research participants must be fully informed of any potential risks and benefits associated with the relevant study [ 8 ]. COPE guidelines provide clear recommendations for publishers, editors, and various research institutes on the topic of publication ethics [ 9 ]. Meanwhile, CARE guidelines are specific to case reports and seek to promote and improve their transparency, accuracy, and usefulness [ 10 ]. The CARE guidelines include a checklist with items such as deidentified patient information, informed consent, and patient perspective on the treatment they received [ 10 ]. While multiple guidelines exist, adherence is not mandatory; 1 study found that out of 50 journals, 76% did not adhere to any guidelines for publication of personal information [ 11 ]. Another study investigating CARE guideline adherence in 36 Indian medical journals found that only a third exhibited average adherence and that overall there was poor reporting of subject-informed consent [ 7 ].
With the growth of online publishing and advancements in technology, case reports from academic journals are widely available as web-based publications, and their reach has expanded to a larger audience [ 4 ]. While increased access to medical case reports may be beneficial, photographs from case reports published in academic journals are now also available in online image search results such as Google Images [ 3 , 4 ]. In such cases, patient photographs circulate beyond the original journal website and can be accessed by anyone using the internet. This raises ethical and legal concerns regarding patients’ informed consent and privacy of health information.
In the original study on this topic, drawing on a sample of case reports with patients who are transgender published between 2008 and 2015, at least 1 patient photograph was available on Google Images for 37% of the medical case reports in the sample [ 3 ]. Curious about whether the results would be the same for a random sample of medical case reports published more recently, the aim of this cross-sectional study was to provide systematic evidence that patient photographs from case reports published in medical journals appear in Google Images search results. Research questions for this study were (1) what percentage of patient medical photographs published in case reports are found in Google Images search results? (2) what is the relationship between open access publication status and image availability? and (3) do the odds of finding patient photographs on third-party websites differ between searches conducted in 2020 and 2022?
Study Design
The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) cross-sectional checklist was used when writing up results [ 12 ] (see Multimedia Appendix 1 ).
Ethical Considerations
Research ethics approval was not required because the data were collected from case reports published in medical journals.
Data Source and Study Population
PubMed includes a diverse range of medical journals and is “the most widely used database with biomedicine-related article abstracts” with over 36 million entries [ 13 ]. The efficient identification of a random sample was facilitated by the ways medical case reports are identified within PubMed. A structured search of PubMed was conducted to identify all indexed medical case reports published within a 1-year period between July 1, 2017, and June 30, 2018 (Search: “2017/07/01” [Date—Publication]: “2018/06/30” (Date—Publication) Filters: Case Reports). The search produced 23,589 results (the search was conducted on August 15, 2018).
Sample Size
To determine sample size, a pilot study was conducted to inform an estimate of effect size and power. All medical case reports indexed in PubMed for the month of February 2018 were identified. This search produced 955 references. Full-text PDFs were retrieved for each reference and the documents were visually checked to see whether each case study included photographic images of patients. Of the 955 case reports published in English in February 2018 and indexed in PubMed, 370 (38.7%) included patient photographs. Based on the original study, it was anticipated that approximately 37% of the case reports with photographs would include at least 1 image found online. Using a 95% Cl and a 4% margin of error, a sample size of 585 was required.
Data Collection and Measures
To identify the random sample for this study, the list of 23,589 case report references was exported from PubMed to Microsoft Excel (Microsoft Corp). A random number generator was used to assign a number to each of the references, and then the list of references was rank-ordered from the smallest to the largest random number. The first 585 references were selected in order, imported into EPPI-Reviewer (EPPI Centre) [ 14 ], and then full-text papers were uploaded for each reference ( Figure 1 ).
For the 585 references, the full text of each case report was examined to determine whether the publication included clinical photographs of patients or not. The photographs from each publication were consecutively numbered on a hard copy, and then the information was entered into a Microsoft Excel spreadsheet, with a unique number for each case report and photograph. A total of 186 case reports included patient photographs, with a total of 598 patient photographs in the sample.
Two categories of data were collected—data at the case report level and data at the image level. At the image level, details were documented related to the specific part of the patient’s body that was photographed (eg, eye, face, or torso); the timing of the photograph (eg, pre- or posttreatment or during surgery); the gender and age of the patient as described in the body of the case report; whether the photograph was in color or not; and whether the authors had attempted to anonymize the photograph using image blurring or bars covering parts of the image. For each case report, 1 member of the team entered data about the images into the Excel spreadsheet. All data were then independently verified by a second team member.
At the case report level, data collection included author information, year of publication, open access status, and availability on Google Scholar, ResearchGate, and PubMed Central. To document the open access format for each of the case reports, 1 member of the team searched for each paper in the open database Unpaywall. The open access status of case reports is classified using colors [ 15 ]. The color classifications are (1) gold—published in an open access journal that is indexed by the Directory of Open Access Journals; (2) green—toll-access on the publisher page, but there is a free copy in an open access repository; (3) hybrid—free under an open license in a toll-access journal; (4) bronze—free to read on the publisher page, but without a clearly identifiable license; and (5) closed—all other papers, including those shared only on academic social networks or Sci-Hub. For the purposes of this analysis, open access included papers categorized as gold, green, hybrid, and bronze.

Google Images Search
To determine whether it was possible to find photographs from the case reports on Google Images, searches were carried out for each of the 186 case reports that included patient photographs. Searches were conducted on a yearly basis from 2019 to 2022, using an approach referred to as algorithmic probing [ 16 ]. This analysis focuses on the results of the most recent searches conducted in 2022.
Three members of the research team (AC, HJ, and ZM) conducted manual searches on Google Images for each paper in the study sample using the same strategy first developed by Marshall et al [ 3 ]. For each reference, the researcher used the title of the case report in quotation marks as the text key. These searches were conducted using a Tor browser, “a proxy that masks the location information and browsing history of the user, allowing for anonymous use of the Internet” [ 17 ]. This browser was used to minimize the influence of Google’s personalization strategies to help prevent results from being skewed by historical searches conducted by team members [ 18 ]. Images of the search result pages were saved in PDF by date. One member of the research team then manually compared the search results in the PDF to the photographs in the published medical case report, circling the matching image using PDF editing software. Google Images search results also include a link directly under the image to the original source of the photograph. In Figure 2 , for example, the first 3 images are from a case report [ 19 ] and include links to BMJ Case Reports, and Europe PMC. The link was extracted for each image and then each source was coded as a journal website, publisher website, research database (eg, Semantic Scholar), research repository (eg, ResearchGate), social media, professional association, or other. A second member of the team verified the results.

Data Management
Data were entered into Microsoft Excel and screened for misentries (eg, spelling errors, empty cells, or shifted cells). The primary outcome variable was the availability of medical photographs on Google Images and was coded as “0” not found and “1” found. Secondary outcome variables included the image source and the availability of images on third-party websites over time. Missing data analyses were performed to screen the data for entry errors.
Statistical Analysis
Sample characteristics were described using means, SDs, quantiles, and frequency distributions. The level of analysis is individual case reports rather than individual photographs. This is necessary for 2 reasons. First, some published figures contain more than 1 patient photograph. For example, a figure may include 4 images of a patient taken from different perspectives. Second, case reports included a range of 1 to 33 patient photographs. The relationship between multiple images found in 1 case report is different from multiple images found in separate case reports, and as a result, each photograph cannot be treated independently.
To better understand whether the characteristics of case reports (such as full-text availability on ResearchGate, PubMed Central, or Google Scholar, and open access status) were related to the availability of medical images on Google Images, chi-square tests and simple logistic regressions were conducted. To test if there is any trend for finding images on third-party websites over different searches over time, generalized estimating equations with a logit link and robust sandwich estimators were used. Odds ratios (OR) with 95% Cl were reported. P ≤.05 was considered significant. Analyses were conducted using SAS (version 9.4; SAS Institute).
Sample Demographics
From the sample of 585 case reports, 186 (31.7%) case reports had at least 1 patient clinical photograph. A total of 598 images were identified in these 186 medical case reports. Individual photographs were coded into five broad categories (1) the specific body part that was photographed; (2) patient sex as identified in the case report; (3) patient age (adult vs child); (4) the timing of the photograph (pretreatment, during surgery, autopsy, etc); and (5) whether the photograph was anonymized or not.
From the 186 case reports with 598 photographs, 309 (51.7%) were photographs of women, 278 (46.2%) were photographs of men, and 3 (0.5%) were photographs of trans women. Information about patient sex or gender was not provided for 8 (1.3%) of the photographs. Patients who were photographed ranged in age from 2 days to 93 years. A total of 412 (68.9%) photographs were taken of adult patients (older than 18 years), and 176 (29.4%) were photographs of infants, children, or teenagers younger than 18 years of age. Information about age was not provided for patients in 10 (1.7%) photographs.
Patient photographs most often included internal organs (eg, endoscopy, laparoscopy, or bronchoscopy; n=151 images, 25.3%). Other common types of photographs included limbs such as legs, arms, hands, or feet (n=109 images, 18.3%), or images of the abdomen or torso (n=53 images, 8.9%). A total of 110 (18.3%) out of 598 photographs included eye, face, and full-body photographs, including 65 (10.9%) that could potentially identify the patient. In terms of the context of when the photograph was taken, 403 (67.4%) were photographs of the patient’s condition pre- or posttreatment whereas 144 (24%) were photographs taken during surgery ( Table 1 ).
| Patient demographic characteristics | Values, n (%) | ||
| Women | 309 (51.7) | ||
| Men | 278 (46.2) | ||
| Trans women | 3 (0.5) | ||
| Unknown | 8 (1.3) | ||
| Adult (18 years or older) | 412 (68.2) | ||
| Infant, child, or teenager (younger than 18 years) | 176 (29.4) | ||
| Unknown | 10 (1.7) | ||
| Internal organs or endoscopy | 151 (25.3) | ||
| Limbs (legs, arms, feet, or hands) | 109 (18.2) | ||
| Mouth | 62 (10.4) | ||
| Torso or abdomen | 53 (8.9) | ||
| Face | 44 (7.4) | ||
| Eyes | 41 (6.9) | ||
| Breasts or chest | 36 (6.0) | ||
| Full body | 25 (4.2) | ||
| Genitals | 22 (3.7) | ||
| Ears | 13 (2.2) | ||
| Head | 13 (2.2) | ||
| Nose | 2 (0.3) | ||
| Photograph of condition | 403 (67.4) | ||
| Presurgery | 7 (1.2) | ||
| During surgery | 144 (24.1) | ||
| Specimen | 39 (6.5) | ||
| Autopsy | 3 (0.5) | ||
| Other | 4 (0.7) | ||
Open Access Status of Case Reports With Medical Images
Of the 186 case reports, 102 (54.8%) were closed access; among the closed-access reports, 66 (65%) case reports had at least 1 image found on Google Images. Of the 83 case reports that were open access, 76 (92%) had at least 1 image found on Google Images. From crude comparisons ( P <.001), it appears that case reports with open access were more likely to have medical images visible as Google Images.
Image Availability
For 76.3% (142/186) of the case reports, at least 1 image was found on Google Images. The odds were higher of finding an image from the case report online if the full-text paper was available on ResearchGate (OR 9.16, 95% Cl 2.71-31.02), PubMed Central (OR 7.90, 95% Cl 2.33-26.77), or Google Scholar (OR 6.07, 95% Cl 2.77-13.29) than if full-text was available solely through an open access journal (OR 5.33, 95% Cl 2.31-12.28), but all factors contribute to increased odds of locating patient images online ( Figure 3 ).

Image Source
To better understand where Google Images is obtaining patient photographs, information about data sources was extracted from the hyperlink under each of the images that were found online. Raw image sources included the journal website, publisher website, research database (eg, Semantic Scholar), research repository (eg, ResearchGate), social media, and professional associations. These were grouped into 2 main categories—journal websites or other websites (any third-party sources). A total of 51.0% of photographs came from the journal website, and 49.0% were from a third-party site. In 2021, 51.1% were from journal websites, and 48.9% from third-party sites. In 2022, the number of images from journal websites increased to 63.4%, while the number from third-party sites was 36.6%.
Trend Over Time
Based on generalized estimating equations, after adjusting for individual study differences, compared with the search in 2020, patient photographs were less likely to be found on third-party websites based on the 2022 search results. Specifically, the odds of finding a patient photograph on a third-party site in 2022 were about 40% less likely, compared with the search done in 2020. This finding was statistically significant with OR 0.61, 95% Cl 0.43-0.87. The likelihood of finding a patient photograph on a third-party website was not significantly different between the search in 2021 and the search in 2020 ( Table 2 ).
| Search | Odd ratio (95% CI) | value |
| 2021 vs 2020 | 1.04 (0.78-1.40) | .77 |
| 2022 vs 2020 | 0.61 (0.43-0.87) | .006 |
Principal Results
The aims of this study were to identify what percentage of patient photographs published in medical case reports were found in Google Images search results, to better understand the relationship between open access publication status and image availability, and to verify whether there is a trend over time for finding patient photographs on third-party websites. Out of the 186 case reports that included clinical photographs, at least 1 photograph from the case report was available on Google Images for 142 (76.3%) references. The odds of finding an image from the case report online were higher if the full-text paper was available on ResearchGate (OR 9.16, 95% CI 2.71-31.02), PubMed Central (OR 7.90, 95% CI 2.33-26.77), or Google Scholar (OR 6.07, 95% CI 2.77-13.29) than if full-text was available solely through an open access journal (OR 5.33, 95% CI 2.31-12.28), but all factors contributed to an increased risk of locating patient images online. This study is the first of its kind to search Google Images for medical photographs from a random sample of case reports; as such there are no studies with which to compare results.
Findings from this study are notably higher than the results from earlier research, where 34 (37%) out of 94 case reports had at least 1 photograph accessible on Google Images [ 3 ]. While the difference in sample population may partially account for the disparity in outcomes, this study identified several additional variables that influenced the availability or unavailability of patient photographs on Google Images. For instance, finding images from the case reports online was more likely if the full-text paper was also available on ResearchGate, PubMed Central, or Google Scholar, compared to case reports solely accessible through open access publications.
To better understand how Google retrieves the images, the image source was recorded for all photographs found on Google Images and these results were compared over a 3-year time period. From 2020 to 2022, there was a notable change in where images were sourced, with a significant decrease in photographs housed on third-party websites such as ResearchGate and Semantic Scholar. This change may be linked to a recent legal judgment where Google was held liable for copyright infringement for displaying content with links to a third-party infringer’s website which was not the original publisher and owner of the copyrighted content [ 20 ].
Limitations
The systematic, documented approach to searching for patient medical photographs on Google Images is a strength of this study. The primary challenge is that Google Images search results are not stable. Although the team attempted to manage as many factors as possible, including using the Tor browser to control for the influence of team member search histories, search results changed. Investigating the same data set yearly for over 3 years, sometimes the photographs were never found, while others were consistently located. The primary findings in this paper are based on the most recent searches in 2022, as the purpose of this study was not to demonstrate the ways search results change over time, but whether the images were found or not. Search results from 2020, 2021, and 2022 are available on request.
A further limitation is that the team did not investigate other image search engines or social media platforms where patient photographs might also appear. While the team was able to provide clear evidence using Google Images it would be an interesting avenue for future research to explore some alternate image search engines and platforms. In addition, the use of the Tor browser to minimize personalization in search results may not completely replicate the typical user experience and may have introduced a form of selection bias.
Conclusions
From a clinical standpoint, the availability of patient photographs on Google Images presents both advantages and risks. Results demonstrated a high proportion of medical photographs from case reports on Google Images. While this concentration allows for wider accessibility and educational benefits, the public availability of these sensitive images online also raises ethical concerns with respect to the privacy of personal health information. Patients should be adequately informed about the possible impacts of providing consent for clinicians to publish their images in medical journals. Even if clinicians seek consent for their publication in case reports, it is not clear whether patients are informed about the possibility of photographs becoming available on Google Images and reaching unintended audiences, including the media and the general public. Similarly, it is not known whether clinicians themselves are aware of these risks. As such, they may not be in a position to ensure informed consent from their patients regarding the potential availability of their clinical images online. A recent content analysis of journal consent forms for the publication of patient photographs found that 55.5% (10/18) of consent forms related to 132 journals mentioned photographs being available to an audience outside of the journal website, but only 16.7% (3/18) addressed the possibility of the patient’s images being linked to journal or publisher social media platforms [ 21 ].
A lack of standardized guidelines poses a challenge to obtaining patient consent for publishing case reports with photographs. In addition to the policy and practice recommendations highlighted in earlier research, current findings underline the need for increased dialogue among academics, patients, governments, and industry. Discussions should focus on improving the consent process and establishing consistent practices and policies for publishing case reports with patient photographs. Study findings indicate that patient photographs are accessible on Google Images, even when published in closed-access case reports. Engagement with Google and other major online image repositories is critical to raise awareness of this issue and to seek input regarding the underlying causes and potential solutions. New policies should be implemented to ensure that patients are protected and that all stakeholders are aware of the risks involved in submitting clinical photographs to online medical journals. Accordingly, the next phase of this study focuses on qualitative interviews with case report authors, journal editors, publishers, and patients. The goal is to identify potential solutions to this complex ethical challenge, including responsive policies that will influence practices across academic publishing to maintain patient privacy.
Acknowledgments
The Natural Sciences and Engineering Research Council of Canada (554764-2021) provided student scholarship funding. The funders had no role in study design, in collection or interpretation of data, in writing the report, or in the decision to submit the paper for publication.
Data Availability
The data sets generated and analyzed during this study are available from the corresponding author on reasonable request.
Authors' Contributions
ZM was responsible for conceptualization (lead), investigation (lead), methodology (equal), supervision (lead), writing—original draft preparation (lead), and writing—review and editing (lead). MB carried out the investigation (equal), project administration (equal), writing—original draft preparation (equal), and writing—review and editing (equal). MW contributed to the formal analysis (lead), methodology (equal), visualization (lead), writing—original draft preparation (equal), and writing—review and editing (supporting). AC did the investigation (equal), writing—original draft preparation (equal), and writing—review and editing (supporting). HJ did the investigation and writing—review and editing (supporting). SA aided in conceptualization (supporting), methodology (equal), writing—original draft preparation (equal), and writing—review and editing (supporting). RP contributed to the investigation (equal), project administration (equal), and writing—review and editing (supporting). VB performed the investigation (equal), project administration (equal), writing—original draft preparation (equal), and writing—review and editing (supporting). VFR did the investigation (equal), project administration (equal), and writing—review and editing (supporting). BC performed the investigation (equal) and writing—review and editing (supporting). LA performed the investigation (equal) and writing—review and editing (supporting). MS contributed to the methodology (equal) and writing—review and editing (supporting). VW aided in conceptualization (supporting), methodology (equal), and writing—review and editing (supporting). FB was involved in conceptualization (supporting) and writing—review and editing (supporting). CK contributed to the conceptualization (supporting) and writing—review and editing (supporting).
Conflicts of Interest
None declared.
Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) cross-sectional checklist.
- Marshall Z, Brunger F, Welch V, Asghari S, Kaposy C. Open availability of patient medical photographs in Google Images search results: cross-sectional study of transgender research. J Med Internet Res. 2018;20(2):e70. [ FREE Full text ] [ CrossRef ] [ Medline ]
- Neely JG, Karni RJ, Nussenbaum B, Paniello RC, Fraley PL, Wang EW, et al. Practical guide to understanding the value of case reports. Otolaryngol Head Neck Surg. 2008;138(3):261-264. [ CrossRef ] [ Medline ]
- Roguljić M, Šimunović D, Peričić TP, Viđak M, Utrobičić A, Marušić M, et al. Publishing identifiable patient photographs in scientific journals: scoping review of policies and practices. J Med Internet Res. 2022;24(8):e37594. [ FREE Full text ] [ CrossRef ] [ Medline ]
- Roguljić M, Peričić TP, Gelemanović A, Jukić A, Šimunović D, Buljan I, et al. What patients, students and doctors think about permission to publish patient photographs in academic journals: a cross-sectional survey in Croatia. Sci Eng Ethics. 2020;26(3):1229-1247. [ CrossRef ] [ Medline ]
- Rees M, Corson SL. Publication of medical case reports and consent. Case Rep Womens Health. 2017;15:A1. [ FREE Full text ] [ CrossRef ] [ Medline ]
- Neavyn M, Murphy C. Coming to a consensus on informed consent for case reports. J Med Toxicol. 2014;10(4):337-339. [ FREE Full text ] [ CrossRef ] [ Medline ]
- Ravi R, Mulkalwar A, Thatte UM, Gogtay NJ. Medical case reports published in PubMed-indexed Indian journals in 2015: adherence to 2013 CARE guidelines. Indian J Med Ethics. 2018;3(3):192-195. [ FREE Full text ] [ CrossRef ] [ Medline ]
- World Medical Association. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310(20):2191-2194. [ CrossRef ] [ Medline ]
- Promoting integrity in research and its publication. Committee on Publication Ethics. URL: https://publicationethics.org/ [accessed 2024-03-22]
- Gagnier JJ, Kienle G, Altman DG, Moher D, Sox H, Riley D, et al. CARE Group. The CARE guidelines: consensus-based clinical case report guideline development. J Clin Epidemiol. 2014;67(1):46-51. [ CrossRef ] [ Medline ]
- Taheri A, Adibi P, Jafari MS, Saeedizadeh M, Rahimi A, Abbasi A. The reporting requirements of case reports and adherence of case report reporting guidelines in medical journals: an analysis of the authors' guide sections. J Med Case Rep. 2023;17(1):2. [ FREE Full text ] [ CrossRef ] [ Medline ]
- von Elm E, Altman DG, Egger M, Pocock SJ, Gøtzsche PC, Vandenbroucke JP, et al. STROBE Initiative. Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. BMJ. 2007;335(7624):806-808. [ FREE Full text ] [ CrossRef ] [ Medline ]
- Novoa J, Chagoyen M, Benito C, Moreno FJ, Pazos F. PMIDigest: interactive review of large collections of PubMed entries to distill relevant information. Genes. 2023;14(4):942. [ FREE Full text ] [ CrossRef ] [ Medline ]
- Thomas J, Graziosi S, Brunton J, Ghouze Z, O'Driscoll P, Bond M, et al. EPPI-Reviewer: advanced software for systematic reviews, maps and evidence synthesis. EPPI Centre. URL: https://eppi.ioe.ac.uk/cms/Default.aspx?tabid=2967 [accessed 2024-04-07]
- Piwowar H, Priem J, Larivière V, Alperin JP, Matthias L, Norlander B, et al. The state of OA: a large-scale analysis of the prevalence and impact of open access articles. PeerJ. 2018;6:e4375. [ FREE Full text ] [ CrossRef ] [ Medline ]
- Rogers R. Algorithmic probing: Prompting offensive Google results and their moderation. Big Data Soc. 2023;10(1). [ CrossRef ]
- Macrina A. Accidental technologist: the Tor browser and intellectual freedom in the digital age. Ref User Serv Q. 2015;54(4):17. [ CrossRef ]
- Bozdag E. Bias in algorithmic filtering and personalization. Ethics Inf Technol. 2013;15(3):209-227. [ CrossRef ]
- van Vonderen JJ, Stol K, Buddingh EP, van der Kaay DCM. Herpes simplex transmission to chest and face through autoinoculation in an infant. BMJ Case Rep. 2017;2017:00. [ FREE Full text ] [ CrossRef ] [ Medline ]
- Bhattacharjee M, Kaposy C, Grossman MR, Marshall Z. Patient photographs on Google Images: a commentary on informed consent, copyright, and privacy laws. Law Innov Technol. 2023;15(2):536-557. [ CrossRef ]
- Asaad L, Bhattacharjee M, Marshall Z. A content analysis of patient medical image consent forms. 2024. Presented at: International Conference on Clinical Ethics and Consultation & Canadian Bioethics Society; May 29-31, 2024; Montreal, Canada. URL: https://event.fourwaves.com/iccec2024/abstracts/d53bdd80-d964-4811-985d-910793a762b1
Abbreviations
| Case Report |
| Committee on Publication Ethics |
| odds ratio |
| Strengthening the Reporting of Observational Studies in Epidemiology |
Edited by T de Azevedo Cardoso; submitted 13.12.23; peer-reviewed by M Roguljić, D Singh; comments to author 08.03.24; revised version received 08.04.24; accepted 24.04.24; published 24.06.24.
©Zack Marshall, Maushumi Bhattacharjee, Meng Wang, Abdul Cadri, Hannah James, Shabnam Asghari, Rene Peltekian, Veronica Benz, Vanessa Finley-Roy, Brynna Childs, Lauren Asaad, Michelle Swab, Vivian Welch, Fern Brunger, Chris Kaposy. Originally published in the Journal of Medical Internet Research (https://www.jmir.org), 24.06.2024.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work, first published in the Journal of Medical Internet Research (ISSN 1438-8871), is properly cited. The complete bibliographic information, a link to the original publication on https://www.jmir.org/, as well as this copyright and license information must be included.
- Skip to main content
- Skip to search
- Skip to footer
Products and Services

Cisco Provider Connectivity Assurance
Gain end-to-end visibility and insights like never before..
Create exceptional digital experiences built on deep network observability and critical network monitoring.
Service assurance that's proactive and precise—a win-win
Boost efficiency and revenue with Cisco Provider Connectivity Assurance (formerly Accedian Skylight), delivering service assurance that continuously monitors and optimizes digital experiences.
Simplified operations and troubleshooting
Gain a single view of granular performance metrics and third-party data to accelerate MTTR.
Continuous optimization of digital experiences
Find transient issues with precise synthetic network and service testing.
Differentiated services, next-level innovation
Premium, SLA-backed services and end-customer portals are just a click away.
Predictive analytics powered by AI
Be proactive, not reactive. Put the power of your multivendor infrastructure to work for a trouble-free network.
Drive additional sales revenue
See more revenue from portal fees and capacity purchases when you give customers deeper visibility and transparency with Cisco Provider Connectivity Assurance.
See what sets Cisco Provider Connectivity Assurance apart
Granular, real-time visibility.
Perform end-to-end and full mesh testing with microsecond precision and make hidden microbursts a thing of the past.
Reliability meets flexibility
Enjoy 99.997% uptime and take full control over network performance for greater reliability.
Optimize your infrastructure
Automatically correlate sensor metrics with third-party sources of performance data to improve ROI and create better digital experiences.
Deliver exceptional services
Ready-made customer portals differentiate services, boost revenue, and enhance satisfaction.
Assuring high-performance critical networks
See how Cisco Provider Connectivity Assurance enables efficient troubleshooting and SLA assurance—all while lowering operational costs.
Cisco Provider Connectivity Assurance use cases
Solve the challenges of fragmented multidomain tools and poor visibility on service quality. Discover how Provider Connectivity Assurance can help you simplify operations.
Real-time SLA visibility
Monitor and police SLAs for accountability, while proactively addressing performance issues.
Critical network monitoring
Improve operational efficiency with proactive assurance and microsecond visibility for faster issue resolution and better experiences.
B2B service differentiation
Create new revenue opportunities to upsell and differentiate your services with multitenant end-customer portals for real-time service-level agreement (SLA) visibility and alerting.
Automated assurance
Detect issues early when you automate assurance for the entire service lifecycle with real-time service visibility and predictive analytics.
Multilayer assurance
Get a single view of performance across multiple network layers, like segment routing and routed optical networking, to reduce tools and drive down MTTR.
Mobile backhaul and edge monitoring
Optimize digital experiences with real-time service visibility, while assuring your end-to-end 5G transport.
Get high-precision service assurance with Cisco Provider Connectivity Assurance
Ai-enabled predictive analytics.
Get an ML-assisted view of your entire network and gain powerful performance insights.
Assurance sensors
Network-wide service performance visibility. Deployable anywhere, at scale.
See why your peers trust Cisco Provider Connectivity Assurance
"we can look at network performance at any level.".
With Cisco Provider Connectivity Assurance, Bouygues Telecom now has a complete "telescopic and microscopic" view of network and service performance in a single tool.
André Ethier, Network Quality Engineer
Bouygues Telecom
"The solution is key in evolving our end-to-end network visibility."
This is extremely powerful in terms of customer experience. It helps to avoid tickets, as customers can see for themselves what happened with their service. This reduces tension and increases customer satisfaction.
Bart Janssens, Senior Specialist, Packet Architecture
Take preventative measures against network degradation.
"With service-centric assurance and granular visibility we can prevent degradations, automate actions for improvements, and better communicate with our customers."
Mahesh Anjan, Senior Product Technology Executive
Better together
Cisco crosswork network automation.
Drive network efficiency and enhance experiences.
Cisco Optics
Plug innovation into your network.
Boost efficiency and service quality
Cisco Provider Connectivity Assurance helps you improve service quality, lower costs, and deliver outstanding user experiences with a single view of service performance across the entire network

IMAGES
VIDEO
COMMENTS
Google Scholar provides a simple way to broadly search for scholarly literature. Search across a wide variety of disciplines and sources: articles, theses, books, abstracts and court opinions.
Access 160+ million publications and connect with 25+ million researchers. Join for free and gain visibility by uploading your research.
Harness the power of visual materials—explore more than 3 million images now on JSTOR. Enhance your scholarly research with underground newspapers, magazines, and journals. Explore collections in the arts, sciences, and literature from the world's leading museums, archives, and scholars. JSTOR is a digital library of academic journals ...
Get 30 days free. 1. Google Scholar. Google Scholar is the clear number one when it comes to academic search engines. It's the power of Google searches applied to research papers and patents. It not only lets you find research papers for all academic disciplines for free but also often provides links to full-text PDF files.
Find the research you need | With 160+ million publications, 1+ million questions, and 25+ million researchers, this is where everyone can access science
Still, Google Books is a great first step to find sources that you can later look for at your campus library. 6. Science.gov. If you're looking for scientific research, Science.gov is a great option. The site provides full-text documents, scientific data, and other resources from federally funded research.
arXiv is a free distribution service and an open-access archive for nearly 2.4 million scholarly articles in the fields of physics, mathematics, computer science, quantitative biology, quantitative finance, statistics, electrical engineering and systems science, and economics.
Make an impact and build your research profile in the open with ScienceOpen. Search and discover relevant research in over 94 million Open Access articles and article records; Share your expertise and get credit by publicly reviewing any article; Publish your poster or preprint and track usage and impact with article- and author-level metrics; Create a topical Collection to advance your ...
Semantic Reader is an augmented reader with the potential to revolutionize scientific reading by making it more accessible and richly contextual. Try it for select papers. Learn More. Semantic Scholar uses groundbreaking AI and engineering to understand the semantics of scientific literature to help Scholars discover relevant research.
Publishing with SpringerOpen makes your work freely available online for everyone, immediately upon publication, and our high-level peer-review and production processes guarantee the quality and reliability of the work. Open access books are published by our Springer imprint. Find the right journal for you. Explore our subject areas.
Free access to millions of research papers for everyone. OA.mg is a search engine for academic papers. Whether you are looking for a specific paper, or for research from a field, or all of an author's works - OA.mg is the place to find it. Universities and researchers funded by the public publish their research in papers, but where do we ...
It is a highly interdisciplinary platform used to search for scholarly articles related to 67 social science topics. SSRN has a variety of research networks for the various topics available through the free scholarly database. The site offers more than 700,000 abstracts and more than 600,000 full-text papers.
1. Google Scholar - The Ultimate Academic Search Engine. Google Scholar is a free-to-use search engine that indexes scholarly articles and other academic materials across disciplines. Users can access a wide range of research papers, articles, theses, and books through Google Scholar.
Get a visual overview of a new academic field. Enter a typical paper and we'll build you a graph of similar papers in the field. Explore and build more graphs for interesting papers that you find - soon you'll have a real, visual understanding of the trends, popular works and dynamics of the field you're interested in.
Work faster and smarter with advanced research discovery tools. Search the full text and citations of our millions of papers. Download groups of related papers to jumpstart your research. Save time with detailed summaries and search alerts. Advanced Search. PDF Packages of 37 papers.
Aggregation plays an increasingly essential role in maximising the long-term benefits of open access, helping to turn the promise of a 'research commons' into a reality. The aggregation services that CORE provides therefore make a very valuable contribution to the evolving open access environment in the UK. Show all.
One of the largest and most authoritative collections of online journals, books, and research resources, covering life, health, social, and physical sciences.
3.3 million articles on ScienceDirect are open access. Articles published open access are peer-reviewed and made freely available for everyone to read, download and reuse in line with the user license displayed on the article. ScienceDirect is the world's leading source for scientific, technical, and medical research.
Best Websites to Download Research Papers. #1. Sci-Hub - Best for Accessing Paywalled Academic Papers. Credits: Armacad. Summary. Unlocks millions of academic articles. Known as the 'Robin Hood' of research. Free and easy to use. Sci-Hub is the defiant maverick of the academic sphere.
About the directory. DOAJ is a unique and extensive index of diverse open access journals from around the world, driven by a growing community, and is committed to ensuring quality content is freely available online for everyone. DOAJ is committed to keeping its services free of charge, including being indexed, and its data freely available.
Free Research Paper Download Websites - 2024. Check the 14 best free websites to download and read research papers listed below: 1. Sci-Hub. Sci-Hub is a website link with over 64.5 million academic papers and articles available for direct download. It bypasses publisher paywalls by allowing access through educational institution proxies.
It works with Google Chrome and Mozilla Firefox, and displays a small padlock icon on the right side of your browser window whenever you visit a page dedicated to a scholarly article. If there's ...
The Open Research Library (ORL) is planned to include all Open Access book content worldwide on one platform for user-friendly discovery, offering a seamless experience navigating more than 20,000 Open Access books.
This paper documents that foreign lobbying influences US government spending. We introduce a comprehensive dataset of over 230,000 date-stamped, in-person meetings between agents representing foreign governments and individual US legislators, state governors, and employees of US executive agencies from 2000 to 2018.
Research questions included the following: (1) what percentage of patient medical photographs published in case reports were found in Google Images search results? ... The odds of finding an image from the case report online were higher if the full-text paper was available on ResearchGate (odds ratio [OR] 9.16, 95% CI 2.71-31.02), PubMed ...
Get a single view of performance across multiple network layers, like segment routing and routed optical networking, to reduce tools and drive down MTTR. ... Get white paper. Take preventative measures against network degradation. "With service-centric assurance and granular visibility we can prevent degradations, automate actions for ...